Logging and Tracking
Contents
The need for logging and tracking
The 'mon' (for monitoring) database contains various tables that hold data to support logging and tracking. Not all these systems are fully developed, but the main elements are in place.
Here 'logging' refers to recording what has happened within the system, during any major operation, and 'tracking' refers to maintaining a record of the state of the system, in particular the status of the various source data packages.
For a study based data sources a 'source data package' is the complete block of data obtained about an individual study, i.e. the study details, the study attribute data, the linked data object details and the object attribute data.
For object based data sources such as PubMed each source data package consists only of the object details and the object attribute data.
In both cases these are distinct packages that they are downloaded and harvested as a unit, even if they are split into different components before being imported and aggregated. There are currently (November 2020) about 600,000 study source data packages tracked by the system, and about 200,000 object source data packages.
Because the 4 major processes (download, harvest, import and aggregation) operate independently of each other, they need to use, and update, a common set of reference data that provides the status of each source data package. This allows, for example, the harvest system to locate the files previously downloaded or created during data download, or identify those that have been downloaded since the last import for any particular source.
The logging systems are required to provider feedback on the system's activity, to check the correct functioning of the system, and to identify apparent errors and anomalies.
System status: The source data tables
The source data tables are used to store the details about the source data packages, and are therefore the chief stores of the system's state. There are two of them - one for studies (sf.source_data_studies) and one for objects (sf.source_data_objects), though they both have exactly the same structure, as listed below.
id integer created as an identity , source_id integer id of the source (= organisation id) , sd_id varchar the object's own id, in the source data (e.g. a registry identifier or PubMed id) , remote_url varchar the URL of its record on the web. This applies even to data that is not collected directly from the web, such as from WHO csv files , last_revised timestampTZ the datetime that the record was last revised, if available , assume_complete boolean indicates if the record is assumed complete (used when no revision date is available) , download_status integer 0 indicates found in a search but not yet (re)downloaded, and 2 indicates downloaded , local_path varchar where the XML file downloaded or created is stored , last_saf_id integer the id of the fetch / search event in which the data package was last downloaded / created , last_downloaded timestampTZ the date-time of that fetch / search , last_harvest_id integer the id of the harvest event in which it was last harvested , last_harvested timestampTZ the date-time of that harvest , last_import_id integer the id of the import event in which it was last imported , last_imported timestampTZ the date-time of that import
Figure 1 provides an image of a few of the rows in the source_data_studies table.
These two tables are accessed and manipulated in the download, harvest and import processes using a data repository class (always called logging_repo) that connects the mon database, and which includes a number of standard (e.g. for fetching or storing individual source date records) or similar functions.
For instance, in the download system, the logging repo contains a function to either create a new source data record (for a newly discovered study or data object) or update the record of an existing record - in particular the revision date, download status and the event id and date-time of the most recent download. In the harvesting system an analogous function updates the id of the last harvest id and the date-time of the harvest, while the import system updates the import event id and the import date-time.
The harvest module's logging_repo also accesses the source data tables to identify the files to be harvested, comparing the dates of last download and import to identify the relevant files. As the code below shows, a SQL statement is constructed using a standard select list (set up in the class's constructor) and then a GetWhereClause() function to generate most of the remaining SQL.
The GetWhereClause uses the harvest type parameter to determine if all the records from a particular source are selected, or just those where the import date-time is null or less than the download date-time.
When large numbers of records are involved a version that adds an offset and limit clause to the SQL statement is available.
...
...(in constructor)
sql_file_select_string = "select id, source_id, sd_id, remote_url, last_revised, ";
sql_file_select_string += " assume_complete, download_status, local_path, last_saf_id, last_downloaded, ";
sql_file_select_string += " last_harvest_id, last_harvested, last_import_id, last_imported ";
...
public IEnumerable<StudyFileRecord> FetchStudyFileRecords(int source_id, int harvest_type_id = 1)
{
string sql_string = sql_file_select_string;
sql_string += " from sf.source_data_studies ";
sql_string += GetWhereClause(source_id, harvest_type_id);
sql_string += " order by local_path";
using (NpgsqlConnection Conn = new NpgsqlConnection(connString))
{
return Conn.Query<StudyFileRecord>(sql_string);
}
}
public IEnumerable<StudyFileRecord> FetchStudyFileRecordsByOffset(int source_id, int offset_num,
int amount, int harvest_type_id = 1)
{
string sql_string = sql_file_select_string;
sql_string += " from sf.source_data_studies ";
sql_string += GetWhereClause(source_id, harvest_type_id);
sql_string += " order by local_path ";
sql_string += " offset " + offset_num.ToString() + " limit " + amount.ToString();
using (NpgsqlConnection Conn = new NpgsqlConnection(connString))
{
return Conn.Query<StudyFileRecord>(sql_string);
}
}
private string GetWhereClause(int source_id, int harvest_type_id)
{
string where_clause = "";
if (harvest_type_id == 1)
{
// Count all files.
where_clause = " where source_id = " + source_id.ToString();
}
else if (harvest_type_id == 2)
{
// Harvest files that have been downloaded since the last import,
// NOTE - not since the last harvest, as multiple harvests may have
// been carried out. A file should be harvested for import if it
// has not yet been imported, or a new download (possible a new version)
// has taken place since the import.
// So files needed where their download date > import date, or they are new
// and therefore have a null import date
where_clause = " where source_id = " + source_id.ToString() +
" and (last_downloaded >= last_imported or last_imported is null) ";
}
where_clause += " and local_path is not null";
return where_clause;
}
The source parameters
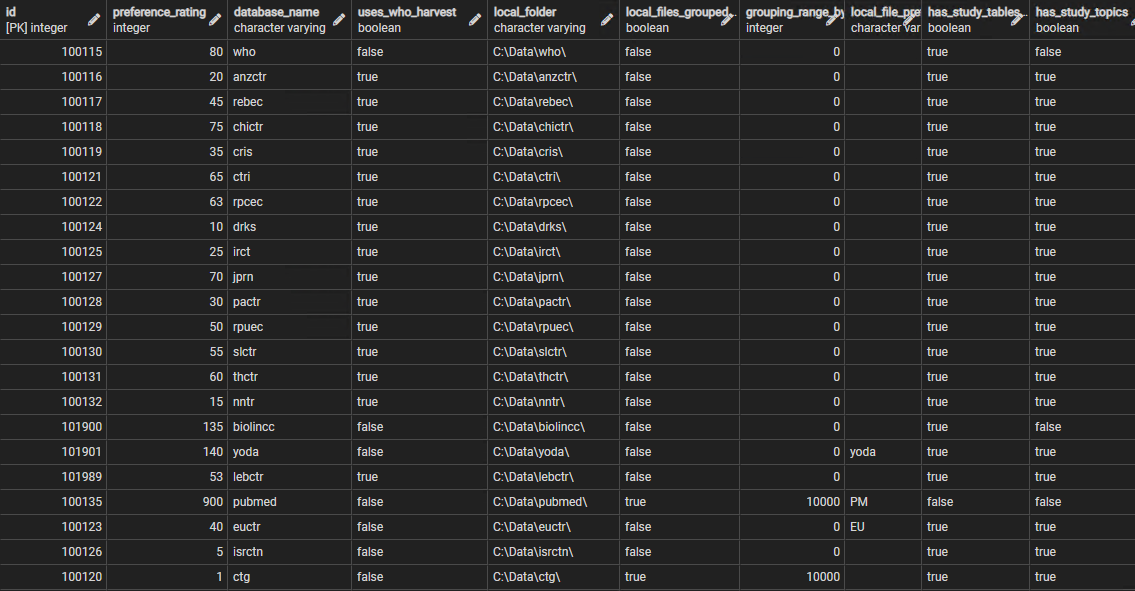
A set of data is required about each data source, and is referenced by all 4 main procedures. The 'source parameter' data indicates which tables are stored in which database, and therefore which which to be included in a table creation, import or aggregation process, and also the local folders where the XML files can be found, and the name of the database. In each case the source data is retrieved into a corresponding Source object, whose properties can then be used to guide the table manipulation processes. For the aggregation process the sources are retrieved as a set, with the process iterating over each source object in turn. The data is held in the sf.source_parameters table, that has the structure below:
id integer (primary key) from ctx.organisations table , preference_rating integer the order in which sources considered , database_name varchar , uses_who_harvest boolean DEFAULT true uses generic WHO data structure , local_folder varchar where XML files stored , local_files_grouped boolean DEFAULT false if sub folders used for local storage , grouping_range_by_id integer DEFAULT 0 quantity in each subfolder , local_file_prefix varchar DEFAULT '' prefix to id if one required , has_study_tables boolean DEFAULT true if includes any study tables , has_study_topics boolean DEFAULT true if includes a study topics table , has_study_features boolean DEFAULT true if includes a study features table , has_study_contributors boolean DEFAULT true if includes a study contributors table , has_study_references boolean DEFAULT false if includes a study references table , has_study_relationships boolean DEFAULT false if includes a study relationships table , has_study_links boolean DEFAULT false if includes a study links table , has_study_ipd_available boolean DEFAULT false if includes a study ipd available table , has_object_datasets boolean DEFAULT false if includes an object datasets table , has_object_dates boolean DEFAULT true if includes an object dates table , has_object_relationships boolean DEFAULT false if includes an object relationships table , has_object_rights boolean DEFAULT false if includes an object rights table , has_object_pubmed_set boolean DEFAULT false if includes the additional objects tables associated with PubMed data
The sources are all included within the contextual organisations table, and their id is derived from their record in that table. The preference rating field is an integer used to order the sources - during aggregation they are processed in the order of this rating (ascending) which indicates the 'preference' for each source to act as the 'prime' data source (see Aggregating_Data). Because several sources share the same WHO dataset structure these can share a common process during download and harvest - the uses_who_harvest field indicates when this is appropriate. In a few cases (ClinicalTrials.gov and PubMed) the number of files is so large that it makes sense to split up the local folder used for storage and group the files, for example dividing them every 10,000 ids, hence the local_files_grouped and grouping_range_by_id fields. In a few cases it is useful to prefix the file name, the id derived from the source, with a small string. The other fields are simple booleans that indicate the presence or absence of tables or groups of tables in the databases corresponding to each source. The defaults simply indicate the most common pattern for each field.
Figure 2 shows a portion of the source parameters table.
The event records
As described above, the source data tables include fields that indicate the Id of the saf (search and fetch), harvest and import event records. The corresponding tables, (sf.saf_events, sf.harvest_events and sf.import_events) are used to provide summary records of each of these processes. for each source (the routines can be called using multiple sources, which are then processed one at a time). Their structures are shown below.
saf_events
id integer (primary key) , source_id integer , type_id integer , time_started timestampTZ , time_ended timestampTZ , num_records_checked integer , num_records_added integer , num_records_downloaded integer , filter_id integer , previous_saf_ids varchar , cut_off_date date, , comments varchar
Here the type_id and filter_id are foreign keys to look up tables with the available types and filters listed. The previous_saf_ids is a list of one or more ids in the same table, when they refer to previous searches (rather than fetches). Depending on the type of search or fetch selected different additional parameters will be required. These are stored in the filter_id, previous_saf_ids and cut_off_date as required.
harvest_events
id integer (primary key) , source_id integer , type_id integer , time_started timestampTZ , time_ended timestampTZ , num_records_available integer , num_records_harvested integer , cut_off_date date , comments varchar
The number of records available refers to the total number of XML fields stored for this source.
import_events
id integer (primary key) , source_id integer , time_started timestampTZ , time_ended timestampTZ , num_new_studies integer , num_edited_studies integer , num_unchanged_studies integer , num_deleted_studies integer , num_new_objects integer , num_edited_objects integer , num_unchanged_objects integer , num_deleted_objects integer , comments varchar
The event records, for search and fetch, harvest and import, are created and filled during each download, harvest and import process respectively. Ids are created manually, in each case, by interrogating the relevant table and incrementing the current max Id. The matching object is created at the beginning of each process, the relevant fields are completed at the end of the process, and the object is then serialised to the database.
Aggregations are slightly different, not least because they involve all the sources in sequence. An aggregation_event record is always created summarising the aggregation process, but additional statistics are available optionally. These include source summaries, which gives the number of records in each table in each source, aggregation summaries, which gives the corresponding numbers of records in each table in the core mdr database, after aggregation, and aggregation object numbers, which gives the total number of each type of data object in the core database.
Logging files
The current feedback produced during the four major processes is reflected to the console, but is also sent to a text log file that is opened at the beginning of each process and closed at its end. This provides details on the timing of each process and the major steps within it, and also records any exception conditions that may have arisen. This is particularly the case for data download, because the remote source may go off line at any time, or block the call - especially if web scraping too quickly - or the internet connection may be interrupted.
It is also possible to create and then serialise in the database an 'extraction note' to record any odd occurrence or a possible problem that will need further investigation. The notes can be one of various types, depending on the nature of the issue being reported, (types are listed in sf.extraction_note_types) and they are stored in the sf.extraction_notes table. Their use is currently relatively limited.