Difference between revisions of "Identifying Links between Studies"
(→Cleaning the 'other Ids') |
|||
| (105 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
| − | ===Introduction=== | + | <p style="color:blue; text-align:right"><small>'''''Last updated: 14/03/2022'''''</small></p> |
| − | One of the issues that has to be tackled during aggregation of data from different sources is the fact that the same study can be found, and in tens of thousands of cases ''is'' found, in more than one study based source, and that it will have a different | + | |
| − | Study titles cannot be relied upon to identify the same study in different source locations. A study title is often expressed slightly differently in different contexts, and cannot in any case be relied upon to be unique (even within the same source). It may be that further research will indicate how titles could be reframed (e.g. to a smaller number of | + | ===Introduction - the problem of duplicates=== |
| + | One of the issues that has to be tackled during aggregation of data from different sources is the fact that the same study can be found, and in tens of thousands of cases ''is'' found, in more than one study based source, and that it will have a different identifier in each source. This is mostly because studies can be registered in more than one trial registry, especially when local regulations mandate a registration for any study carried out within a particular country or region. This is the case with the EU, which insists all trials involving medicinal products that are run in the EU must be registered in the EUCTR. About a third of these studies, however, are also registered in other registries, especially Clinicaltrials.gov. In addition, within a data repository, studies will usually be referenced by a local id rather than a pre-existing registry id.<br/> | ||
| + | Study titles cannot be relied upon to identify the same study in different source locations. A study title is often expressed slightly differently in different contexts, and cannot in any case be relied upon to be unique (even within the same source). It may be that further research will indicate how titles could be reframed (e.g. to a smaller number of lexemes, expressed in a fixed order) to allow duplicate entries to be discovered using text, but for the moment the only easy way of identifying duplicate studies is by using the 'other identifiers' material found in the source data. Almost all sources contain this material, which includes 'other trial registry' ids, (i.e. other than that used in the source registry entry), as well as ids assigned by the sponsor, funder or, sometimes, a regulatory authority.<br/> | ||
These 'other registry ids' can be used to build up a table of study-study links which can then be used during the aggregation process to identify when duplicate studies are being added. In fact the creation of this table is always the first stage of any aggregation. This page describes this process in detail.<br/> | These 'other registry ids' can be used to build up a table of study-study links which can then be used during the aggregation process to identify when duplicate studies are being added. In fact the creation of this table is always the first stage of any aggregation. This page describes this process in detail.<br/> | ||
| − | + | This page only describes how '''duplicate study entries''' are managed. Duplication between data objects is much rarer, with one exception - the PubMed citations for journal papers. These can also be found multiple times in the source data, both within a single source and across multiple sources. The particular complications in processing PubMed data are described in '''[[Processing PubMed Data]]'''. The way in which duplications of data objects that are not PubMed citations are managed is described more fully in '''[[Aggregating Data]]'''. | |
| + | |||
===The Preferred Source concept=== | ===The Preferred Source concept=== | ||
If details about a study and its attributes can be found in more than one data source, the obvious question is how should this data be merged in the final aggregated MDR database? In fact there are several aspects to this question: | If details about a study and its attributes can be found in more than one data source, the obvious question is how should this data be merged in the final aggregated MDR database? In fact there are several aspects to this question: | ||
| − | * How should 'single-occurrence' details about the study be merged? (i.e. the data points that appear in the study record itself, such as study display title, study type and status, enrolment target, min and max ages etc.) | + | * How should 'single-occurrence' details about the study be merged? (i.e. the data points that appear once, in the study record itself, such as study display title, study type and status, enrolment target, min and max ages etc.) |
* How should study attributes (identifiers, titles, contributors, topics etc.) be merged? | * How should study attributes (identifiers, titles, contributors, topics etc.) be merged? | ||
| − | * How should data objects in different sources (trial registry entries, data and document references) be merged? | + | * How should the data objects linked to the same study in different sources (trial registry entries, data and document references) be merged? |
* How should data object attributes be merged? | * How should data object attributes be merged? | ||
| − | The first question is probably the most difficult. If the system allowed these basic study parameters to be merged from a variety of sources the problem of | + | The first question is probably the most difficult. If the system allowed these basic study parameters to be merged from a variety of sources the problem of precedence immediately arises: how could the system 'know' which source to use for each parameter, if they were available in both? For instance how would the system select the enrolment target from one repository rather than another? (There is no guarantee, unfortunately, that the same values will be listed). If the data is edited in one source but not another, should the most recent data always take precedence over the old? (it may be less complete than what is already there). <br/> |
Because of these issues it was decided to | Because of these issues it was decided to | ||
| − | * Only take the study single occurrence data items from a single data source | + | * Only take the study single occurrence data items from a ''single'' data source |
* Use the idea of 'preferred sources' to order the precedence of sources in a consistent fashion. | * Use the idea of 'preferred sources' to order the precedence of sources in a consistent fashion. | ||
| − | + | Each data source is therefore given a number that indicate its place in this 'order of preference' - the exact number is not important, only the place it gives to the data source when they are listed, in ascending order, using this parameter. The number is stored within the '''sf.source_parameters''' table in the '''mon''' database, and is available as a property of the Source objects when they are retrieved from this table. '''''During the aggregation process, the data sources are processed in order, most preferred first, down to the 'least preferred''''''. In general, more preferred sources have richer or at least more consistent data than less preferred sources. The most preferred source is ClincalTrials.gov, followed (in broad terms) by registries that are extracted individually, then the various trial registries using the WHO dataset, and then data repositories (e.g. Yoda, BioLINCC), and finally object based data sources (PubMed). <br/> | |
| − | Although it cannot be guaranteed in every case, this usually means that the richer data is added to the system first, and that data coming from later, less preferred sources augments rather than replaces it. In general (and as described in more detail in '''[[Aggregating Data]]'''), during aggregation the core databases are recreated and then completely rebuilt by adding data from each source in turn | + | Although it cannot be guaranteed in every case, this usually means that the richer data is added to the system first, and that data coming from later, less preferred sources augments rather than replaces it. In general (and as described in more detail in '''[[Aggregating Data]]'''), during aggregation the core databases are recreated and then completely rebuilt by adding data from each source in turn. If a study does not already exist in the core system when it is 'presented' from a data source database, then it, its attributes, its associated data objects and their attributes are all added during the aggregation process. But if a study is added that already exists in the core system, because it has been added from a 'more preferred' source earlier in the process: |
| − | If a study does not already exist in the core system when it is 'presented' from a data source database, then it, its attributes, its associated data objects and their attributes are all added during the aggregation process. | + | * The single-occurrence study details from the new source are ignored |
| − | But if a study is added that already exists in the core system, because it has been added from a 'more preferred' source earlier in the process: | + | * Study attributes are only added if they are clearly different from any that have already been added |
| − | * The single-occurrence study details are ignored | + | * Data objects are added (unless, very rarely, they can be seen to be already in the system) |
| − | * Study attributes are only added if they are | + | * Data object attributes are added, for all the data object records that are added. |
| − | * Data objects are added (unless, very rarely, they can be seen to be already | + | The Preferred Source idea also plays a central role in establishing and using the study-study link data. If a study has multiple source (registry / repository) identifiers in the system, then in general each of those identifiers, other than the 'most preferred', will need to be linked to the 'most preferred' identifier (i.e. the one used by the 'most preferred' source). This is the role of the '''nk.study_study_links table'''. When additional 'less preferred' data for a study is added to the system, the first most-preferred instance of the study is guaranteed to already be present in the central mdr study tables, because of the ordering of the addition. The links table is used to match and compare the new data with the existing set, and so determine how the data will be added. How the '''nk.study_study_links table''' is constructed is explained in detail below. |
| − | * Data object attributes are added | + | |
| + | ===Initial Links Data Collection=== | ||
| + | The initial step is to collect in all the 'other identifier' information from the source databases. <br/> | ||
| + | Two temporary tables are established in the links (nk) schema of the mdr database. One simply collects the link data found, and the second is used to hold it once it has been ordered properly. | ||
| + | <div style="font-family: monospace; font-size: 13px" > | ||
| + | <pre> | ||
| + | public void SetUpTempLinkCollectorTable() | ||
| + | { | ||
| + | using (var conn = new NpgsqlConnection(connString)) | ||
| + | { | ||
| + | string sql_string = @"DROP TABLE IF EXISTS nk.temp_study_links_collector; | ||
| + | CREATE TABLE nk.temp_study_links_collector( | ||
| + | source_1 int | ||
| + | , sd_sid_1 varchar | ||
| + | , sd_sid_2 varchar | ||
| + | , source_2 int) "; | ||
| + | conn.Execute(sql_string); | ||
| + | } | ||
| + | } | ||
| + | |||
| + | public void SetUpTempLinkSortedTable() | ||
| + | { | ||
| + | using (var conn = new NpgsqlConnection(connString)) | ||
| + | { | ||
| + | string sql_string = @"DROP TABLE IF EXISTS nk.temp_study_links_sorted; | ||
| + | CREATE TABLE nk.temp_study_links_sorted( | ||
| + | source_id int | ||
| + | , sd_sid varchar | ||
| + | , preferred_sd_sid varchar | ||
| + | , preferred_source_id int) "; | ||
| + | conn.Execute(sql_string); | ||
| + | } | ||
| + | } | ||
| + | </pre> | ||
| + | </div> | ||
| + | The system then iterates through all the source, interrogating each of the associated databases in turn to bring back the 'other identifier' data, as sd_sid_2, and the id of the organisation assigning the identifier as source_2. That is matched against the source organisation (source_1) and the study's identifier sd_sid (sd_sid_1). The data is pre-filtered to include only identifiers associated with trial registries (and not those assigned from sponsors or funders, etc.). Repository data sources are included in this process but the repository identifiers themselves are not returned - so Yoda and BioLINCC (for example) identifiers appear as sd_sid_1, with a matching sd_sid_2 from a trial registry, but never as sd_sid_2. The data is brought back as an IEnumerable collection of StudyLink objects, which is then immediately stored in the temp_study_links_collector table using the PostgreSQLCopyHelper nuget package, which allows quick and easy storage of batches of data in one call. As the iteration across the sources proceeds, so this table gradually grows with the records found each time. | ||
| + | <div style="font-family: monospace; font-size: 13px" > | ||
| + | <pre> | ||
| + | foreach (Source s in sources) | ||
| + | { | ||
| + | // Fetch the study-study links and store them | ||
| + | // in the Collector table | ||
| + | IEnumerable<StudyLink> links = slh.FetchLinks(s.id, s.database_name); | ||
| + | slh.StoreLinksInTempTable(CopyHelpers.links_helper, links); | ||
| + | } | ||
| + | |||
| + | public IEnumerable<StudyLink> FetchLinks(int source_id, string database_name) | ||
| + | { | ||
| + | string conn_string = repo.GetConnString(database_name); | ||
| + | using (var conn = new NpgsqlConnection(conn_string)) | ||
| + | { | ||
| + | string sql_string = @"select " + source_id.ToString() + @" as source_1, | ||
| + | sd_sid as sd_sid_1, | ||
| + | identifier_value as sd_sid_2, identifier_org_id as source_2 | ||
| + | from ad.study_identifiers | ||
| + | where identifier_type_id = 11 | ||
| + | and identifier_org_id > 100115 | ||
| + | and (identifier_org_id < 100133 or identifier_org_id = 101989) | ||
| + | and identifier_org_id <> " + source_id.ToString(); | ||
| + | return conn.Query<StudyLink>(sql_string); | ||
| + | } | ||
| + | } | ||
| − | === | + | public ulong StoreLinksInTempTable(PostgreSQLCopyHelper<StudyLink> copyHelper, IEnumerable<StudyLink> entities) |
| + | { | ||
| + | using (var conn = new NpgsqlConnection(connString)) | ||
| + | { | ||
| + | conn.Open(); | ||
| + | return copyHelper.SaveAll(conn, entities); | ||
| + | } | ||
| + | } | ||
| + | </pre> | ||
| + | </div> | ||
| + | |||
| + | ===Cleaning the 'other Ids'=== | ||
| + | One of the difficulties in the process is that the other identifier data is rarely constrained in the source systems - it can therefore sometimes be poorly formatted or otherwise mis-entered. It is necessary to try and clean some of the data before proceeding further, using a succession of SQL statements - a few of which are shown below. In the examples shown, the SQL a) deletes WHO Universal trial numbers incorrectly entered as trial registry Ids, b) replaces 'n dashes' with ordinary hyphens, making later processing easier, c) updates entries indicating an Australian ACTRN identifier, that are not prefixed by 'ACTRN', and d) removes the redundant 'Chinese Clinical Trial Register' statement from Ids from that registry which include it. These are, however, just 4 examples from a total of over 30 'cleaning' functions. | ||
| + | <div style="font-family: monospace; font-size: 13px" > | ||
| + | <pre> | ||
| + | public void TidyIds1() | ||
| + | { | ||
| + | string sql_string = ""; | ||
| + | using (var conn = new NpgsqlConnection(connString)) | ||
| + | { | ||
| + | sql_string = @"DELETE from nk.temp_study_links_collector | ||
| + | where sd_sid_2 ilike 'U1111%' or sd_sid_2 ilike 'UTRN%'"; | ||
| + | conn.Execute(sql_string); | ||
| + | |||
| + | // replace n dashes | ||
| + | sql_string = @"UPDATE nk.temp_study_links_collector | ||
| + | set sd_sid_2 = replace(sd_sid_2, '–', '-');"; | ||
| + | conn.Execute(sql_string); | ||
| + | |||
| + | sql_string = @"UPDATE nk.temp_study_links_collector | ||
| + | SET sd_sid_2 = 'ACTRN' || sd_sid_2 | ||
| + | WHERE source_2 = 100116 | ||
| + | and length(sd_sid_2) = 14"; | ||
| + | conn.Execute(sql_string); | ||
| + | |||
| + | sql_string = @"UPDATE nk.temp_study_links_collector | ||
| + | set sd_sid_2 = replace(sd_sid_2, 'Chinese Clinical Trial Register', '') | ||
| + | where source_2 = 100118;"; | ||
| + | conn.Execute(sql_string); | ||
| + | |||
| + | ... | ||
| + | ... | ||
| + | </pre> | ||
| + | </div> | ||
| + | Once the data is cleaned it needs to be ordered properly, so that preferred and non-preferred ids are clearly identified. This is done by adding the collected data to the nk.temp_study_links_sorted table using two insert statements. The first takes the data that already has the less preferred source/id (i.e. has the higher preference number) as the first two fields, and puts this data straight into the the 'sorted' table. The second takes those where the less preferred source/id are in the last two fields and swaps them round before inserting the data. | ||
| + | <div style="font-family: monospace; font-size: 13px" > | ||
| + | <pre> | ||
| + | public void TransferLinksToSortedTable() | ||
| + | { | ||
| + | using (var conn = new NpgsqlConnection(_connString)) | ||
| + | { | ||
| + | // needs to be done twice to keep the ordering of sources correct | ||
| + | // A lower rating means 'more preferred' - i.e. should be used in preference | ||
| + | // Therefore lower rated source data should be in the 'preferred' fields | ||
| + | // and higher rated data should be on the left hand side | ||
| + | |||
| + | // Original data matches what is required | ||
| + | |||
| + | string sql_string = @"INSERT INTO nk.temp_study_links_sorted( | ||
| + | source_id, sd_sid, preferred_sd_sid, preferred_source_id) | ||
| + | SELECT t.source_1, t.sd_sid_1, t.sd_sid_2, t.source_2 | ||
| + | FROM nk.temp_study_links_collector t | ||
| + | inner join nk.temp_preferences r1 | ||
| + | on t.source_1 = r1.id | ||
| + | inner join nk.temp_preferences r2 | ||
| + | on t.source_2 = r2.id | ||
| + | WHERE r1.preference_rating > r2.preference_rating"; | ||
| + | |||
| + | int res1 = conn.Execute(sql_string); | ||
| + | |||
| + | // Original data is the opposite of what is required - therefore switch | ||
| + | |||
| + | sql_string = @"INSERT INTO nk.temp_study_links_sorted( | ||
| + | source_id, sd_sid, preferred_sd_sid, preferred_source_id) | ||
| + | SELECT t.source_2, t.sd_sid_2, t.sd_sid_1, t.source_1 | ||
| + | FROM nk.temp_study_links_collector t | ||
| + | inner join nk.temp_preferences r1 | ||
| + | on t.source_1 = r1.id | ||
| + | inner join nk.temp_preferences r2 | ||
| + | on t.source_2 = r2.id | ||
| + | WHERE r1.preference_rating < r2.preference_rating"; | ||
| + | |||
| + | int res2 = conn.Execute(sql_string); | ||
| + | _logger.Information((res1 + res2).ToString() + " total study-study links found in source data"); | ||
| + | } | ||
| + | } | ||
| + | </pre> | ||
| + | </div> | ||
| + | The sorted table will obviously have many duplicates - data that was entered originally as both source_id_1/sd_sid_1 <=> source_id_2/sd_sid_2, ''and'' source_id_2/sd_sid_2 <=> source_id_1/sd_sid_1, will now appear as two identical records: source_id/sd_sid <=> pref_source_id/pref_sd_sid. It is therefore necessary to generate a distinct version of the links data. The '''nk.temp_distinct_links''' table is therefore created, containing all the distinct links between studies, with both the more 'preferred' and the less 'preferred' versions of each pair identified. Note that a boolean field called ''valid'' is included in the table, with a default value of true. The '''nk.temp_distinct_links''' table is used as the maim links table during both the final stages of data cleaning and the next stages of processing. | ||
| + | <div style="font-family: monospace; font-size: 13px" > | ||
| + | <pre> | ||
| + | public void CreateDistinctSourceLinksTable() | ||
| + | { | ||
| + | // The nk.temp_study_links_sorted table will have | ||
| + | // many duplicates... create a distinct version of the data | ||
| + | |||
| + | using (var conn = new NpgsqlConnection(_connString)) | ||
| + | { | ||
| + | string sql_string = @"DROP TABLE IF EXISTS nk.temp_distinct_links; | ||
| + | CREATE TABLE nk.temp_distinct_links | ||
| + | as SELECT distinct source_id, sd_sid, | ||
| + | preferred_sd_sid, preferred_source_id, true as valid | ||
| + | FROM nk.temp_study_links_sorted"; | ||
| + | |||
| + | conn.Execute(sql_string); | ||
| + | |||
| + | sql_string = @"SELECT COUNT(*) FROM nk.temp_distinct_links"; | ||
| + | int res = conn.ExecuteScalar<int>(sql_string); | ||
| + | _logger.Information(res.ToString() + " distinct study-study links found"); | ||
| + | } | ||
| + | } | ||
| + | </pre> | ||
| + | </div> | ||
| + | Even though the 'other ids' are now sorted and (mostly) properly formatted they may not all be valid. There may have been errors during data entry, or some other reason why the registry Ids as entered do not correspond to a real study in the system. These invalid ids have to be removed, or the system will try to link data to non-existent studies, causing a variety of problems during the aggregation process. The system therefore loops through each of the sources with study tables, and uses a call to the source database to retrieve a list of the current study ids for that source. These are all guaranteed to be 'real' ids as they have been harvested directly from the sources as the registry ids. They are also the most up to date set of study ids known to the system, being a function of the most recent import processes. | ||
| + | <div style="font-family: monospace; font-size: 13px" > | ||
| + | <pre> | ||
| + | foreach (Source source in sources) | ||
| + | { | ||
| + | ... | ||
| + | if (source.has_study_tables) | ||
| + | { | ||
| + | string source_conn_string = _credentials.GetConnectionString(source.database_name, _testing); | ||
| + | slh.ObtainStudyIds(source.id, source_conn_string, CopyHelpers.studyids_checker); | ||
| + | slh.CheckIdsAgainstSourceStudyIds(source.id); | ||
| + | } | ||
| + | } | ||
| + | slh.DeleteInvalidLinks(); | ||
| + | |||
| + | public void ObtainStudyIds(int source_id, string source_conn_string, PostgreSQLCopyHelper<IdChecker> copyHelper) | ||
| + | { | ||
| + | using (var conn = new NpgsqlConnection(_connString)) | ||
| + | { | ||
| + | string sql_string = @"DROP TABLE IF EXISTS nk.temp_id_checker; | ||
| + | CREATE TABLE nk.temp_id_checker | ||
| + | (sd_sid VARCHAR) "; | ||
| + | conn.Execute(sql_string); | ||
| + | } | ||
| + | |||
| + | IEnumerable<IdChecker> Ids; | ||
| + | using (var conn = new NpgsqlConnection(source_conn_string)) | ||
| + | { | ||
| + | string sql_string = @"select sd_sid | ||
| + | from ad.studies"; | ||
| + | Ids = conn.Query<IdChecker>(sql_string); | ||
| + | } | ||
| + | using (var conn = new NpgsqlConnection(_connString)) | ||
| + | { | ||
| + | conn.Open(); | ||
| + | copyHelper.SaveAll(conn, Ids); | ||
| + | } | ||
| + | ... | ||
| + | } | ||
| + | </pre> | ||
| + | </div> | ||
| + | Once the study ids have been collected the IDs in the '''nk.temp_distinct_links''' table can be checked against them. Both sides (preferred and non-preferred) must be checked separately. If a link contains an invalid study id then it is marked as a invalid. At the end of the process, after all sources have been checked, all the invalid records are deleted. In most cases the proportion of invalid records is very small. In fact only about 50 records are identified (from 32,000 plus) as invalid across all the sources, with one marked exception. That exception is the European Clinical Trials Registry (EU CTR). In this case over 5000 Ids, often called EUDRACT numbers, listed in other trial registries as an 'other registry id', do ''not'' correspond to any entry in the EU CTR itself. Checking the relevant titles / key words in several cases indicate that this is not a data entry error issue - the trials simply do not exist in the EU CTR, under any Id. This may be a function of sponsors seeking an EU CTR Id in anticipation of running a study within the EU, or to allow possible expansion to an EU country, but without proceeding to do so. | ||
| + | <div style="font-family: monospace; font-size: 13px" > | ||
| + | <pre> | ||
| + | public void CheckIdsAgainstSourceStudyIds(int source_id) | ||
| + | { | ||
| + | using (var conn = new NpgsqlConnection(_connString)) | ||
| + | { | ||
| + | string sql_string = @"UPDATE nk.temp_distinct_links t | ||
| + | SET valid = false | ||
| + | FROM | ||
| + | (SELECT k.sd_sid as sd_sid | ||
| + | FROM nk.temp_distinct_links k | ||
| + | LEFT JOIN nk.temp_id_checker s | ||
| + | ON k.sd_sid = s.sd_sid | ||
| + | WHERE k.source_id = " + source_id.ToString() + @" | ||
| + | AND s.sd_sid is null) invalids | ||
| + | where t.sd_sid = invalids.sd_sid"; | ||
| + | int res1 = conn.Execute(sql_string); | ||
| + | _logger.Information(res1.ToString() + " set to invalid on sd_sid"); | ||
| + | sql_string = @"UPDATE nk.temp_distinct_links t | ||
| + | SET valid = false | ||
| + | FROM | ||
| + | (SELECT k.preferred_sd_sid | ||
| + | FROM nk.temp_distinct_links k | ||
| + | LEFT JOIN nk.temp_id_checker s | ||
| + | ON k.preferred_sd_sid = s.sd_sid | ||
| + | WHERE k.preferred_source_id = " + source_id.ToString() + @" | ||
| + | AND s.sd_sid is null) invalids | ||
| + | where t.preferred_sd_sid = invalids.preferred_sd_sid"; | ||
| + | int res2 = conn.Execute(sql_string); | ||
| + | _logger.Information(res2.ToString() + " set to invalid on preferred_sd_sid"); | ||
| + | } | ||
| + | } | ||
| + | </pre> | ||
| + | </div> | ||
| + | The last stage of the process carries out the deletion of the invalid records, drops a temporary table and counts the number of records available for further processing. Importantly it also drops the 'valid' column from the table. That is because it would not be consistentlky updated in later processing, and so would cause the several later calls to 'select distinct' to not function properly. It is therefore simpler to remove it. | ||
| + | <div style="font-family: monospace; font-size: 13px" > | ||
| + | <pre> | ||
| + | public void DeleteInvalidLinks() | ||
| + | { | ||
| + | using (var conn = new NpgsqlConnection(_connString)) | ||
| + | { | ||
| + | string sql_string = @"DELETE | ||
| + | FROM nk.temp_distinct_links | ||
| + | WHERE valid = false"; | ||
| + | int res = conn.Execute(sql_string); | ||
| + | _logger.Information(res.ToString() + " study-study links deleted because of an invalid study id"); | ||
| − | = | + | sql_string = @"DROP TABLE IF EXISTS nk.temp_id_checker;"; |
| + | conn.Execute(sql_string); | ||
| + | sql_string = @"ALTER TABLE nk.temp_distinct_links DROP COLUMN valid;"; | ||
| + | conn.Execute(sql_string); | ||
| + | sql_string = @"SELECT COUNT(*) FROM nk.temp_distinct_links"; | ||
| + | res = conn.ExecuteScalar<int>(sql_string); | ||
| + | _logger.Information(res.ToString() + " distinct study-study links remaining"); | ||
| + | } | ||
| + | } | ||
| + | </pre> | ||
| + | </div> | ||
===Exclusion of One-to-Many links=== | ===Exclusion of One-to-Many links=== | ||
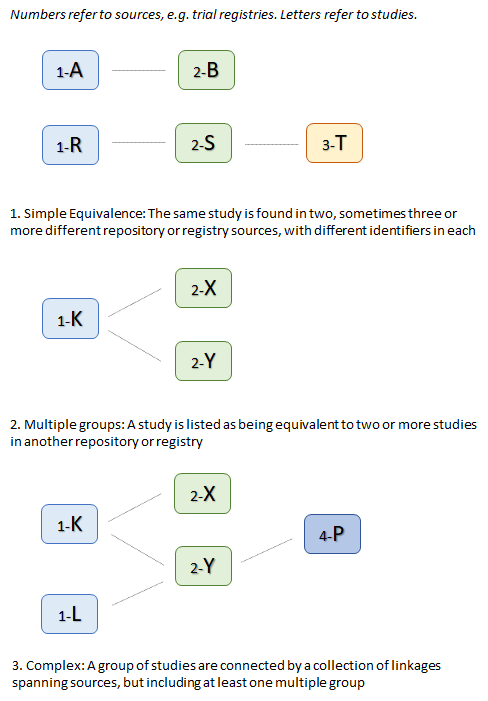
| + | An additional issue is that not all of the links identified between studies are 'simple' one-to-one links. In some cases they are one-to-many. In other words, rather than having the same study listed under different identifiers in different contexts, (scenario 1 in figure 1 below) we have a single study in one source listed as two or more studies in another source (scenario 2). In these circumstances the MDR does not attempt to merge the study data. The system cannot know how data objects and attributes listed on the 'One' side could be allocated to the studies listed on the 'Many' side, so it keeps all studies that are in this sort of relationship as individual studies.<br/> | ||
| + | In fact the situation can be more complex - for 30 - 40 groups of studies the relationships are many-to-many. In this case a study may be listed as equivalent to two or more studies in another registry or source, one of which is itself listed as two or more studies (see scenario 3 in figure 1). Usually one of the second set will be the original study, but others are also included. In such case a a group of 4,5 or more (in one case 10!) are listed as equivalent studies in some way. The groupings appear to be a mix of exact equivalents, registered in muiltiple locations, and closely related studies that are listed as equivalent but are not in fact exactly the same (e.g. they may be a follow up study). | ||
| + | [[File:study_link_types.png|center||]] | ||
| + | <p style="text-align:center"><b>Figure 1: Types of linkage between studies</b></p> | ||
| + | Although the MDR does not attempt to merge the data from one-to-many or many-to-many groupings, it does record the fact that these relationships exist, as a 'study_relationship' record. While some study relationships are between studies listed in the same data source, the ones considered here clearly cut across different sources. The task is therefore to identify this form of linkage and to extract it as study relationship data, removing it from the larger group of 'linked studies'. The first stage is to establish a temporary table that can hold identified study groups, and then identify the groups within '''nk.temp_distinct_links'''. A 'group' in this context is two or more studies that are all in the same data source (have the same source id) that are all matched to a particular study. The matching could be at either the preferred or less preferred side of the data so two insert statements are required. Note that at this stage it is just the 'grouping' study and source that are identified (the 'One' side) - not the members of that group (the 'Many' side). | ||
| + | <div style="font-family: monospace; font-size: 13px" > | ||
| + | <pre> | ||
| + | private void Identify1ToNGroupedStudies() | ||
| + | { | ||
| + | string sql_string; | ||
| + | |||
| + | using (var conn = new NpgsqlConnection(_connString)) | ||
| + | { | ||
| + | // create a table that takes the rows that involve | ||
| + | // studies with multiple matches in the same source | ||
| + | |||
| + | sql_string = @"DROP TABLE IF EXISTS nk.temp_linked_studies; | ||
| + | CREATE TABLE nk.temp_linked_studies( | ||
| + | group_source_id int | ||
| + | , group_sd_sid varchar | ||
| + | , member_sd_sid varchar | ||
| + | , member_source_id int | ||
| + | , source_side varchar | ||
| + | , complex int DEFAULT 0 | ||
| + | )"; | ||
| + | |||
| + | conn.Execute(sql_string); | ||
| + | |||
| + | // The source_id side is the group and the preferred side | ||
| + | // is comprised of the grouped studies. | ||
| + | |||
| + | sql_string = @"INSERT into nk.temp_linked_studies | ||
| + | (group_source_id, group_sd_sid, | ||
| + | member_source_id, member_sd_sid, | ||
| + | source_side) | ||
| + | SELECT k.source_id, k.sd_sid, | ||
| + | k.preferred_source_id, k.preferred_sd_sid, 'L' | ||
| + | from nk.temp_distinct_links k | ||
| + | inner join | ||
| + | (SELECT source_id, sd_sid | ||
| + | FROM nk.temp_distinct_links | ||
| + | group by source_id, sd_sid, preferred_source_id | ||
| + | HAVING count(sd_sid) > 1) lhs_groups | ||
| + | ON k.source_id = lhs_groups.source_id | ||
| + | AND k.sd_sid = lhs_groups.sd_sid"; | ||
| + | |||
| + | conn.Execute(sql_string); | ||
| + | |||
| + | // The preferred_source_id side is the group and the | ||
| + | // non-prefrred side is comprised of grouped studies | ||
| + | |||
| + | sql_string = @"INSERT into nk.temp_linked_studies | ||
| + | (group_source_id, group_sd_sid, | ||
| + | member_source_id, member_sd_sid, | ||
| + | source_side) | ||
| + | SELECT k.preferred_source_id, k.preferred_sd_sid, | ||
| + | k.source_id, k.sd_sid, 'R' from | ||
| + | nk.temp_distinct_links k | ||
| + | inner join | ||
| + | (SELECT preferred_source_id, preferred_sd_sid | ||
| + | FROM nk.temp_distinct_links | ||
| + | group by preferred_source_id, preferred_sd_sid, source_id | ||
| + | HAVING count(preferred_sd_sid) > 1) rhs_groups | ||
| + | ON k.preferred_source_id = rhs_groups.preferred_source_id | ||
| + | AND k.preferred_sd_sid = rhs_groups.preferred_sd_sid"; | ||
| + | |||
| + | conn.Execute(sql_string); | ||
| + | } | ||
| + | } | ||
| + | </pre> | ||
| + | </div> | ||
| + | The temp_linked_studies table now holds all the grouped studies, with the grouping source/studies on the left hand side, and the grouped source/studies on the right. The next stage is to differntiate the studies that belong to the many-to-many relationships, by flagging studies in groups which have members that themselves group other studies. This is done by adding an integer 'complex' flag to the relevant records, first to those involved in the top level group, and then to those in the 'child' group. | ||
| + | <div style="font-family: monospace; font-size: 13px" > | ||
| + | <pre> | ||
| + | private void IdentifyNToNGroupedStudies() | ||
| + | { | ||
| + | string sql_string; | ||
| + | using (var conn = new NpgsqlConnection(_connString)) | ||
| + | { | ||
| + | // ... | ||
| + | |||
| + | sql_string = @"Update nk.temp_linked_studies ks1 | ||
| + | SET complex = ks1.complex + 2 | ||
| + | FROM nk.temp_linked_studies ks2 | ||
| + | WHERE ks1.member_source_id = ks2.group_source_id | ||
| + | AND ks1.member_sd_sid = ks2.group_sd_sid | ||
| + | AND ks1.source_side <> ks2.source_side"; | ||
| + | |||
| + | conn.Execute(sql_string); | ||
| + | |||
| + | sql_string = @"Update nk.temp_linked_studies ks1 | ||
| + | SET complex = ks1.complex + 4 | ||
| + | FROM nk.temp_linked_studies ks2 | ||
| + | WHERE ks1.group_source_id = ks2.group_source_id | ||
| + | AND ks1.group_sd_sid = ks2.group_sd_sid | ||
| + | AND ks2.complex = 2"; | ||
| + | |||
| + | conn.Execute(sql_string); | ||
| + | } | ||
| + | |||
| + | </pre> | ||
| + | </div> | ||
| + | The temp_linked_data can now be used to create linked study group records with the correct relationship codes. For the simpler set of multiple records (complex score = 0) this is straightforward and carried out with the code below. Two complementary pairs of such relationships are possible: | ||
| + | * 25 Includes target as one of a group of non-registered studies: This study includes<the target study>.That study is not registered independently, but instead shares this registry entry with one or more other non-registered studies. | ||
| + | * 26 Non registered but included within a registered study group: This study is registered as <the target study>, along with one or more other studies that share the same registry entry and id. | ||
| + | * 28 Includes target as one of a group of registered studies: This study includes <the target study>, which is registered elsewhere along with one or more other registered studies, forming a group that collectively equates to this study. | ||
| + | * 29 Registered and is included elsewhere in group: This study is also registered, along with one or more other studies that together form an equivalent group, as <the target study>. | ||
| + | BioLINCC (id = 101900) and Yoda (id = 101901) are the only two non registry study sources at the moment, and this is reflected in the SQL below. The relationship codes for these data repositories are different than for the trial registries. | ||
| + | <div style="font-family: monospace; font-size: 13px" > | ||
| + | <pre> | ||
| + | private void Extract1ToNGroupedStudies() | ||
| + | { | ||
| + | string sql_string; | ||
| + | using (var conn = new NpgsqlConnection(_connString)) | ||
| + | { | ||
| + | // ... | ||
| + | |||
| + | sql_string = @"INSERT INTO nk.linked_study_groups | ||
| + | (source_id, sd_sid, relationship_id, | ||
| + | target_sd_sid, target_source_id) | ||
| + | select group_source_id, group_sd_sid, | ||
| + | case when group_source_id = 101900 | ||
| + | or group_source_id = 101901 then 25 | ||
| + | else 28 end, | ||
| + | member_sd_sid, member_source_id | ||
| + | from nk.temp_linked_studies | ||
| + | where complex = 0;"; | ||
| + | |||
| + | int res1 = conn.Execute(sql_string); | ||
| + | _logger.Information(res1.ToString() + " relationship records added as part of 1 to n study relationships"); | ||
| + | |||
| + | sql_string = @"INSERT INTO nk.linked_study_groups | ||
| + | (source_id, sd_sid, relationship_id, | ||
| + | target_sd_sid, target_source_id) | ||
| + | select member_source_id, member_sd_sid, | ||
| + | case when member_source_id = 101900 | ||
| + | or member_source_id = 101901 then 26 | ||
| + | else 29 end, | ||
| + | group_sd_sid, group_source_id | ||
| + | from nk.temp_linked_studies | ||
| + | where complex = 0;"; | ||
| + | |||
| + | int res2 = conn.Execute(sql_string); | ||
| + | _logger.Information(res2.ToString() + " relationship records added as part of n to 1 study relationships"); | ||
| + | } | ||
| + | } | ||
| + | </pre> | ||
| + | </div> | ||
| + | For the 'complex' groups, although only a relatrively small number are involved, the processing required is quite extensive. The initial task is to extract the groups and their members into a uniform format. The code below does that, using a series of union statements to bring together the various grouping studies and their members. (As usual in line comments have been removed for brevity). | ||
| + | <div style="font-family: monospace; font-size: 13px" > | ||
| + | <pre> | ||
| + | private void ExtractNToNGroupedStudyData() | ||
| + | { | ||
| + | string sql_string; | ||
| + | |||
| + | using (var conn = new NpgsqlConnection(_connString)) | ||
| + | { | ||
| + | sql_string = @"drop table if exists nk.temp_multi_links; | ||
| + | create table nk.temp_multi_links as | ||
| + | select s1.group_source_id as s1groupsource, s1.group_sd_sid as s1groupsdsid, | ||
| + | s1.member_source_id as s1membersource, s1.member_sd_sid as s1membersdsid, | ||
| + | s1.complex as s1complex, | ||
| + | s2.group_source_id as s2groupsource, s2.group_sd_sid as s2groupsdsid, | ||
| + | s2.member_source_id as s2membersource, s2.member_sd_sid as s2membersdsid, | ||
| + | s2.complex as s2complex | ||
| + | from nk.temp_linked_studies s1 | ||
| + | left join | ||
| + | nk.temp_linked_studies s2 | ||
| + | on s1.member_sd_sid = s2.group_sd_sid | ||
| + | where s1.complex > 0 | ||
| + | order by s1.group_sd_sid, s1.complex desc, s2.complex desc "; | ||
| + | |||
| + | conn.Execute(sql_string); | ||
| + | |||
| + | sql_string = @"drop table if exists nk.temp_list_studies; | ||
| + | create table nk.temp_list_studies as | ||
| + | select * from | ||
| + | (select s1groupsource as src_id, s1groupsdsid as sid_id, s1groupsource as sourceid, s1groupsdsid as sdsid | ||
| + | from nk.temp_multi_links | ||
| + | union | ||
| + | select s1groupsource, s1groupsdsid, s1membersource, s1membersdsid | ||
| + | from nk.temp_multi_links | ||
| + | union | ||
| + | select s1groupsource, s1groupsdsid, s2groupsource, s2groupsdsid | ||
| + | from nk.temp_multi_links | ||
| + | where s2groupsource is not null | ||
| + | union | ||
| + | select s1groupsource, s1groupsdsid, s2membersource, s2membersdsid | ||
| + | from nk.temp_multi_links | ||
| + | where s2groupsource is not null) tot | ||
| + | order by src_id, sid_id, sourceid, sdsid; "; | ||
| + | conn.Execute(sql_string); | ||
| + | } | ||
| + | } | ||
| + | </pre> | ||
| + | </div> | ||
| + | The difficulty is that the source data will have multiple copies of the same or similar groups, but they will be collected under different 'grouping studies'. This is because the original data is read in multiple different ways, so that a different member of the group will be the 'top' grouping study in each case. It is necessary to rationalise the data so that there is only a single set of group records. The code below does this. It tests the members of each group to see if any of them correspond to a ''previous'' grouping study. If a match is found all the members of that group are added to the previous study's group rather than being left as an independent group. In that way the original 70+ groups are reduced to 30+. There will be mnany duplicates within each group, so these are removed using a select distinct call in SQL. | ||
| + | <div style="font-family: monospace; font-size: 13px" > | ||
| + | <pre> | ||
| + | private void RationaliseNToNGroupedStudyData() | ||
| + | { | ||
| + | string sql_string; | ||
| + | using (var conn = new NpgsqlConnection(_connString)) | ||
| + | { | ||
| + | // First task is to get all entries in related key study lists to the same (first of the group) key study list | ||
| + | sql_string = @"select distinct src_id, sid_id, false from nk.temp_list_studies order by src_id, sid_id"; | ||
| + | IEnumerable<ComplexKeyStudy> key_studies = conn.Query<ComplexKeyStudy>(sql_string); | ||
| + | int current_key_src_id = 0, new_key_src_id = 0, current_src_id = 0; | ||
| + | string current_key_sid_id = "", new_key_sid_id = "", current_sid_id = ""; | ||
| + | |||
| + | foreach (ComplexKeyStudy ks in key_studies) | ||
| + | { | ||
| + | // ... | ||
| + | current_key_src_id = ks.src_id; | ||
| + | current_key_sid_id = ks.sid_id; | ||
| + | |||
| + | sql_string = @"select sourceid, sdsid | ||
| + | from nk.temp_list_studies | ||
| + | where src_id = " + current_key_src_id.ToString() + @" | ||
| + | and sid_id = '" + current_key_sid_id + @"' | ||
| + | order by sourceid, sdsid"; | ||
| + | IEnumerable<ComplexStudy> studies = conn.Query<ComplexStudy>(sql_string); | ||
| + | bool match_found = false; | ||
| + | |||
| + | foreach (ComplexStudy s in studies) | ||
| + | { | ||
| + | if (!match_found) // if still looking | ||
| + | { | ||
| + | current_src_id = s.sourceid; | ||
| + | current_sid_id = s.sdsid; | ||
| + | |||
| + | foreach (ComplexKeyStudy kks in key_studies) | ||
| + | { | ||
| + | if (kks.considered == true) // for the previous 'considered' key studies only... | ||
| + | { | ||
| + | // does the current study entry matrch any of those? | ||
| + | // If so record the details and break out of the loop | ||
| + | |||
| + | if (kks.src_id == current_src_id && kks.sid_id == current_sid_id) | ||
| + | { | ||
| + | match_found = true; | ||
| + | new_key_src_id = kks.src_id; | ||
| + | new_key_sid_id = kks.sid_id; | ||
| + | break; | ||
| + | } | ||
| + | } | ||
| + | } | ||
| + | } | ||
| + | } | ||
| + | |||
| + | if (match_found) | ||
| + | { | ||
| + | // this is a partial or complete repeat of the existing group | ||
| + | // update the table | ||
| + | sql_string = @"UPDATE nk.temp_list_studies | ||
| + | set src_id = " + new_key_src_id.ToString() + @" | ||
| + | , sid_id = '" + new_key_sid_id + @"' | ||
| + | where src_id = " + current_key_src_id.ToString() + @" | ||
| + | and sid_id = '" + current_key_sid_id + "'"; | ||
| + | |||
| + | conn.Execute(sql_string); | ||
| + | } | ||
| + | |||
| + | // if no match and is a 'new group' - can be left as it is | ||
| + | ks.considered = true; // signal this key study as 'considered' | ||
| + | } | ||
| + | |||
| + | // Lots of duplicates now so use SQL to selct distinct | ||
| + | sql_string = @"DROP TABLE if exists nk.temp_list_studies2; | ||
| + | CREATE TABLE nk.temp_list_studies2 as | ||
| + | SELECT distinct * from nk.temp_list_studies order by src_id, sid_id, sourceid, sdsid; | ||
| + | DROP TABLE if exists nk.temp_list_studies; | ||
| + | ALTER TABLE nk.temp_list_studies2 rename to temp_list_studies "; | ||
| + | conn.Execute(sql_string); | ||
| + | } | ||
| + | } | ||
| + | </pre> | ||
| + | </div> | ||
| + | It is now relatively straightforward to loop through the rationalised table and create study relationship records. In this case the relationship is type 30: 'Is grouped with, and is the same or similar to <target study>'. Each study has to be given this relationship record with every other member of its group, and then the reverse record, with the role of source and target reversed, is also constructed. A group of 6 related studies will therefore generate 30 relationship records (2 x (5 + 4 + 3 + 2 + 1)). | ||
| + | <div style="font-family: monospace; font-size: 13px" > | ||
| + | <pre> | ||
| + | private void StoreNToNGroupedStudyData() | ||
| + | { | ||
| + | string sql_string; | ||
| + | using (var conn = new NpgsqlConnection(_connString)) | ||
| + | { | ||
| + | sql_string = @"select distinct src_id, sid_id from nk.temp_list_studies order by src_id, sid_id"; | ||
| + | IEnumerable<ComplexKeyStudy> key_studies = conn.Query<ComplexKeyStudy>(sql_string); | ||
| + | |||
| + | List<ComplexLink> links = new List<ComplexLink>(); | ||
| + | foreach (ComplexKeyStudy ks in key_studies) | ||
| + | { | ||
| + | // get the studies related to the current key study | ||
| + | sql_string = @"select sourceid, sdsid | ||
| + | from nk.temp_list_studies | ||
| + | where src_id = " + ks.src_id.ToString() + @" | ||
| + | and sid_id = '" + ks.sid_id + @"' | ||
| + | order by sourceid, sdsid"; | ||
| + | |||
| + | List<ComplexStudy> studies = conn.Query<ComplexStudy>(sql_string).AsList<ComplexStudy>(); | ||
| + | int studynum = studies.Count; | ||
| + | for (int i = 0; i < studynum - 1; i++) | ||
| + | { | ||
| + | ComplexStudy s = studies[i]; | ||
| + | for (int j = i + 1; j < studynum; j++) | ||
| + | { | ||
| + | ComplexStudy t = studies[j]; | ||
| + | links.Add(new ComplexLink(s.sourceid, s.sdsid, 30, t.sourceid, t.sdsid)); | ||
| + | links.Add(new ComplexLink(t.sourceid, t.sdsid, 30, s.sourceid, s.sdsid)); | ||
| + | } | ||
| + | } | ||
| + | } | ||
| + | |||
| + | // store list of relationships, between all possible pairs | ||
| + | conn.Open(); | ||
| + | CopyHelpers.complex_links_helper.SaveAll(conn, links); | ||
| + | _logger.Information(links.Count.ToString() + " relationship records added as part of n to n study relationships"); | ||
| + | } | ||
| + | } | ||
| + | </pre> | ||
| + | </div> | ||
| + | Finally, it is necessary to remove the grouped study data from the main set of '''nk.temp_distinct_links''', as shown below, and drop the temporary tables that have been used. | ||
| + | <div style="font-family: monospace; font-size: 13px" > | ||
| + | <pre> | ||
| + | private void DeleteGroupedStudyLinkRecords() | ||
| + | { | ||
| + | string sql_string; | ||
| + | using (var conn = new NpgsqlConnection(_connString)) | ||
| + | { | ||
| + | sql_string = @"DELETE FROM nk.temp_distinct_links k | ||
| + | USING nk.temp_linked_studies g | ||
| + | WHERE k.source_id = g.group_source_id | ||
| + | and k.sd_sid = g.group_sd_sid | ||
| + | and k.preferred_source_id = g.member_source_id | ||
| + | and g.source_side = 'L';"; | ||
| + | int res1 = conn.Execute(sql_string); | ||
| + | |||
| + | sql_string = @"DELETE FROM nk.temp_distinct_links k | ||
| + | USING nk.temp_linked_studies g | ||
| + | WHERE k.preferred_source_id = g.group_source_id | ||
| + | and k.preferred_sd_sid = g.group_sd_sid | ||
| + | and k.source_id = g.member_source_id | ||
| + | and g.source_side = 'R';"; | ||
| + | int res2 = conn.Execute(sql_string); | ||
| + | _logger.Information((res1 + res2).ToString() + " study-study links extracted as grouped (1 to n, n to n) records"); | ||
| + | |||
| + | sql_string = @"DROP TABLE IF EXISTS nk.temp_grouping_studies; | ||
| + | DROP TABLE IF EXISTS nk.temp_list_studies; | ||
| + | DROP TABLE IF EXISTS nk.temp_multi_links; | ||
| + | DROP TABLE IF EXISTS nk.temp_linked_studies"; | ||
| + | conn.Execute(sql_string); | ||
| + | |||
| + | sql_string = @"SELECT COUNT(*) FROM nk.temp_distinct_links"; | ||
| + | int res = conn.ExecuteScalar<int>(sql_string); | ||
| + | _logger.Information(res.ToString() + " distinct study-study links remaining"); | ||
| + | } | ||
| + | } | ||
| + | </pre> | ||
| + | </div> | ||
| + | |||
| + | ===Cascading Links=== | ||
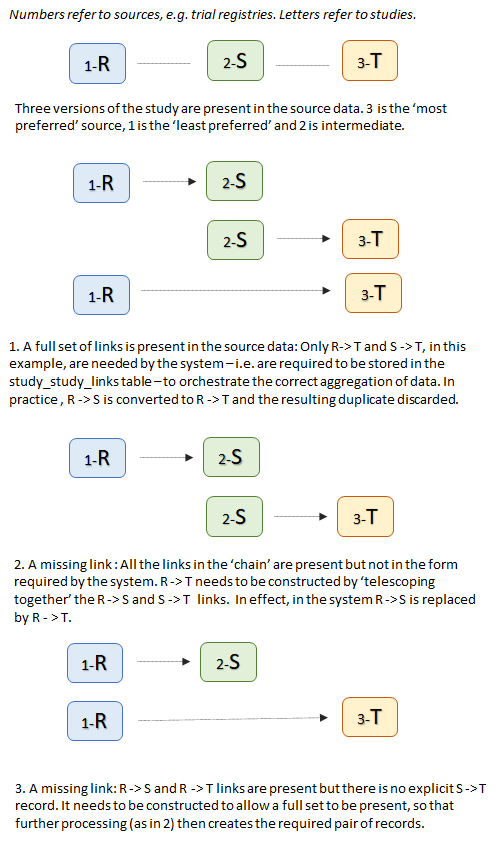
| + | The linked study data requires further processing, however, to ensure that it is complete and can be used for the aggregation process. Two additional problems can arise when a study is found in '''''three or more''''' sources, and need to be resolved. These occur because there is no guarantee that all possible links, or the 'correct' links, as shown in scenario 1 of figure 2, were in the original data.<br/> | ||
| + | The first problem is that although the links table currently identifies the more 'preferred' of two linked study ids, it does not necessarily identify the ''most'' preferred when there are three or more links. For instance a study could have identifiers in sources A, B and C, where A is less preferred than B, and B less preferred than C. In the source data there are links between A and B, and B and C, but no link exists between A and C, so the data shows A -> B and B -> C. For the aggregation process to work properly the data has to show A -> C and B -> C, i.e. all links should be to the ''most preferred'' id. The B -> C link is fine, but the A -> B link has to be 'telescoped' with the 'B -> C' link, and replaced by the resultant A -> C link. This is represented by scenario 2 of figure 2, (though using R, S and T). The system carries out this telescoping process whether or not the A -> C link was originally present in the data. If it was it is duplicated and the duplicate is simply removed. If it was not then the required link record is generated.<br/> | ||
| + | The second problem is that to carry out the telescoping behaviour described above requires the intermediate link (B -> C in the example here) to be present. That link may be missing, as in scenario 3 of figure 2, where there is R -> S and R -> T but no S -> T. The system must therefore add these 'missing links' ''before'' it can carry out the telescoping process and ensure that all links point to the most preferred version of the study data. | ||
| + | [[File:study_link_repair.png|center||]] | ||
| + | <p style="text-align:center"><b>Figure 2: Repairing the linkage between 3 or more studies</b></p> | ||
| + | The first step in adding the missing intermediate links is to identify studies that have more than one 'preferred' option - these will always link across to more than one preferred source (e.g. R -> S, R -> T) because the grouped studies (linking to the same source / study) have already been removed. This list of studies (in the example all the studies of type 'R') is inserted into '''temp_studies_with_multiple_links'''. The process then creates the '''temp_missing_links''' table, adding the right hand side preferred fields, by joining this table with the records in the main links table '''temp_distinct_links''', at the same time incorporating the preference ratings of the sources involved. '''temp_missing_links''' therefore holds the 'records of interest' from the original links table - all the R -> S, R -> T pairs - together with the preference ratings for all the Rs, Ss and Ts. | ||
| + | <div style="font-family: monospace; font-size: 13px" > | ||
| + | <pre> | ||
| + | public void AddMissingLinks() | ||
| + | { | ||
| + | string sql_string; | ||
| + | using (var conn = new NpgsqlConnection(_connString)) | ||
| + | { | ||
| + | // ... | ||
| + | sql_string = @"DROP TABLE IF EXISTS nk.temp_studies_with_multiple_links; | ||
| + | CREATE TABLE nk.temp_studies_with_multiple_links | ||
| + | as SELECT source_id, sd_sid | ||
| + | from nk.temp_distinct_links | ||
| + | group by source_id, sd_sid | ||
| + | having count(distinct preferred_source_id) > 1;"; | ||
| + | conn.Execute(sql_string); | ||
| + | |||
| + | sql_string = @"DROP TABLE IF EXISTS nk.temp_missing_links; | ||
| + | CREATE TABLE nk.temp_missing_links as | ||
| + | select k.source_id, r1.preference_rating as source_rating, | ||
| + | k.sd_sid, k.preferred_source_id, r2.preference_rating, k.preferred_sd_sid | ||
| + | from nk.temp_distinct_links k | ||
| + | inner join nk.temp_studies_with_multiple_links m | ||
| + | on k.source_id = m.source_id | ||
| + | and k.sd_sid = m.sd_sid | ||
| + | inner join nk.temp_preferences r1 | ||
| + | on k.source_id = r1.id | ||
| + | inner join nk.temp_preferences r2 | ||
| + | on k.preferred_source_id = r2.id | ||
| + | order by k.source_id, k.sd_sid, preferred_source_id;"; | ||
| + | conn.Execute(sql_string); | ||
| + | |||
| + | ... | ||
| + | ... | ||
| + | </pre> | ||
| + | </div> | ||
| + | Now, within the same function, a further temp table is created ('''temp_new_links''') to construct the missing links between the E and F studies. This table ('''temp_new_links''') has 6 fields - two initially populated with the source id / sd_sid, to identify the source records in '''nk.temp_missing_links''', and the next pair that have the source / sd_sid pair that has the least preferred rating, i.e. is the study that will need to be on the left hand side in the new link (E in the example above). Place holders are put in for the new_preferred_source and new_preferred_sd_sid fields. Then the table is updated, but this time with the source / sd_sid pair that has the more preferred rating, i.e. is the study that will need to be on the right hand side in the new link (F in the example above) going into the new_preferred_ fields. Thus, if '''nk.temp_missing_links''' includes the fields<br/> | ||
| + | D-source id D-sd_sid D-preference E-source id E-sd_sid E-preference<br/> | ||
| + | D-source id D-sd_sid D-preference F-source id F-sd_sid F-preference<br/> | ||
| + | The fields in '''temp_new_links''' will initially be:<br/> | ||
| + | D-source id D-sd_sid E-source id E-sd_sid 0 '--'<br/> | ||
| + | D-source id D-sd_sid E-source id E-sd_sid 0 '--'<br/> | ||
| + | and are then updated to:<br/> | ||
| + | D-source id D-sd_sid E-source id E-sd_sid F-source F-sd_sid<br/> | ||
| + | D-source id D-sd_sid E-source id E-sd_sid F-source F-sd_sid<br/> | ||
| + | The four right hand fields now represent the missing (E -> F) links that were required and as such can be inserted into the temp_distinct_links table (as a distinct selection, as in some cases multiple records are created. Of course in some cases the E -> F links may have already existed, as well as the D -> E, D -> F links. The additional record is then redundant, and is removed by making reselecting the link records as a distincct set, but the process has to be carried out on the assumption that the E -> F type links are missing. Finally, the temporary tables are dropped. | ||
| + | <div style="font-family: monospace; font-size: 13px" > | ||
| + | <pre> | ||
| + | public void AddMissingLinks() | ||
| + | { | ||
| + | ... | ||
| + | ... | ||
| + | |||
| + | sql_string = @"DROP TABLE IF EXISTS nk.temp_new_links; | ||
| + | CREATE TABLE nk.temp_new_links as | ||
| + | select m.source_id, m.sd_sid, m.preferred_source_id as new_source_id, | ||
| + | m.preferred_sd_sid as new_sd_sid, 0 as new_preferred_source, '' as new_preferred_sd_sid from | ||
| + | nk.temp_missing_links m | ||
| + | inner join | ||
| + | (select source_id, sd_sid, min(preference_rating) as min_rating | ||
| + | from nk.temp_missing_links | ||
| + | group by source_id, sd_sid) mins | ||
| + | on m.source_id = mins.source_id | ||
| + | and m.sd_sid = mins.sd_sid | ||
| + | and m.preference_rating <> mins.min_rating | ||
| + | order by source_id, sd_sid;"; | ||
| + | conn.Execute(sql_string); | ||
| + | |||
| + | sql_string = @"UPDATE nk.temp_new_links k | ||
| + | SET new_preferred_source = min_set.preferred_source_id | ||
| + | , new_preferred_sd_sid = min_set.preferred_sd_sid | ||
| + | FROM | ||
| + | (select m.* from | ||
| + | nk.temp_missing_links m | ||
| + | INNER JOIN | ||
| + | (select source_id, sd_sid, min(preference_rating) as min_rating | ||
| + | from nk.temp_missing_links | ||
| + | group by source_id, sd_sid) mins | ||
| + | on m.source_id = mins.source_id | ||
| + | and m.sd_sid = mins.sd_sid | ||
| + | and m.preference_rating = mins.min_rating) min_set | ||
| + | WHERE k.source_id = min_set.source_id | ||
| + | AND k.sd_sid = min_set.sd_sid;"; | ||
| + | conn.Execute(sql_string); | ||
| + | |||
| + | sql_string = @"INSERT INTO nk.temp_distinct_links | ||
| + | (source_id, sd_sid, preferred_sd_sid, preferred_source_id) | ||
| + | SELECT distinct new_source_id, new_sd_sid, new_preferred_sd_sid, new_preferred_source from | ||
| + | nk.temp_new_links;"; | ||
| + | int res = conn.Execute(sql_string); | ||
| + | _logger.Information(res.ToString() + " new study-study links added to complete linkage chains"); | ||
| + | |||
| + | // drop the temp tables | ||
| + | sql_string = @"DROP TABLE IF EXISTS nk.temp_missing_links; | ||
| + | DROP TABLE IF EXISTS nk.temp_studies_with_multiple_links; | ||
| + | DROP TABLE IF EXISTS nk.temp_new_links;"; | ||
| + | conn.Execute(sql_string); | ||
| + | } | ||
| + | MakeLinksDistinct(); | ||
| + | } | ||
| + | |||
| + | </pre> | ||
| + | </div> | ||
| + | Having filled in any 'missing links' the system can safely tackle the telescoping of links. In this scenario, (both A -> B and B -> C exist but not A -> C) the same study (B in this case) will appear in both the left hand side 'less preferred' columns in the temp_distinct_links table ''and'' in the more preferred right hand side. The requirement is to replace the B on the right hand side with the 'true' most preferred study id, which is C. A self join on the temp_distinct_links table, linking studies on both 'sides' of the table, can be used to first see if there are any studies that are both 'less' and 'more' preferred, and then to make the switch, replacing the preferred side data on the right of the table with the 'most preferred' values. In the example, A -> B is linked to B -> C by the join, and A -> B becomes A -> C after the update, 'telescoping' the links together.<br/> | ||
| + | Because a few studies are registered 4 or even more times the process needs to be repeated until no further studies are found which exist in both the less and more preferred columns. A while loop is therefore used to repeat the action as often as is necessary - usually twice.<br/> | ||
| + | In some cases the telescoped link may have already been present - i.e. A -> B, B -> C and A -> C were all there. In these cases the process described above (still necessary to remove the A -> B link) will result in duplicates. Any duplicates therefore have to be removed at the end of the process. This is done by a select distinct. The code below, as usual, has in-line comments reemoved for brevity. | ||
| + | <div style="font-family: monospace; font-size: 13px" > | ||
| + | <pre> | ||
| + | public void CascadeLinks() | ||
| + | { | ||
| + | using (var conn = new NpgsqlConnection(_connString)) | ||
| + | { | ||
| + | string sql_string; | ||
| + | int match_number = 500; // arbitrary start number | ||
| + | while (match_number > 0) | ||
| + | { | ||
| + | sql_string = @"SELECT count(*) | ||
| + | FROM nk.temp_distinct_links t1 | ||
| + | inner join nk.temp_distinct_links t2 | ||
| + | on t1.preferred_source_id = t2.source_id | ||
| + | and t1.preferred_sd_sid = t2.sd_sid"; | ||
| + | |||
| + | match_number = conn.ExecuteScalar<int>(sql_string); | ||
| + | _logger.Information(match_number.ToString() + " cascading study-study links found, to 'telescope'"); | ||
| + | |||
| + | if (match_number > 0) | ||
| + | { | ||
| + | // do the update | ||
| + | sql_string = @"UPDATE nk.temp_distinct_links t1 | ||
| + | SET preferred_source_id = t2.preferred_source_id, | ||
| + | preferred_sd_sid = t2.preferred_sd_sid | ||
| + | FROM nk.temp_distinct_links t2 | ||
| + | WHERE t1.preferred_source_id = t2.source_id | ||
| + | AND t1.preferred_sd_sid = t2.sd_sid"; | ||
| + | |||
| + | conn.Execute(sql_string); | ||
| + | } | ||
| + | } | ||
| + | } | ||
| + | |||
| + | MakeLinksDistinct(); | ||
| + | } | ||
| + | </pre> | ||
| + | </div> | ||
| + | The processing described above, of first adding any missing links and then telescoping the links together, can lead to a small number of new 'groupings' appearing, when more than one study in a single source is linked to a single study elsewhere. The system therefore re-runs the 'exclusion of one-many' routines, to remove these from the '''nk.temp_distinct_links''' table. | ||
| + | ===Updating links with Study_Id === | ||
| + | The data is then transferred from the temporary table to the study_study_links table - a table that is created anew at the beginning of each aggregation porcess. Again a select distinct is used, as a final check to ensure that any duplicates are removed. <br/> | ||
| + | <div style="font-family: monospace; font-size: 13px" > | ||
| + | <pre> | ||
| + | public void TransferNewLinksToDataTable() | ||
| + | { | ||
| + | // A distinct selection is required because the most recent | ||
| + | // link cascade may have generated duplicates | ||
| + | |||
| + | using (var conn = new NpgsqlConnection(connString)) | ||
| + | { | ||
| + | string sql_string = @"Insert into nk.study_study_links | ||
| + | (source_id, sd_sid, preferred_sd_sid, preferred_source_id) | ||
| + | select distinct source_id, sd_sid, preferred_sd_sid, preferred_source_id | ||
| + | from nk.temp_distinct_links"; | ||
| + | |||
| + | conn.Execute(sql_string); | ||
| + | } | ||
| + | } | ||
| + | </pre> | ||
| + | </div> | ||
| + | The study links table is then updated with the '''study Id''' to be used for each non-preferred / preferred pair, as described below, and is then available to play two critical roles during the aggregation process. | ||
| + | * To ensure that a study Id is allocated consistently across all aggregations, and | ||
| + | * To check if a study is a 'non-preferred' version of a study that has been added earlier, and thus modify the details of the data aggregation process | ||
| + | These two tasks are distinct because a study's non-preferred entry may be added to the MDR before its ''preferred'' set of data, for example the EU CTR entry may be added a week before the same study appears in ClinicalTrials.gov. In that example, the persistent study Id within the system (the ECRIN ID) will be allocated when the EUCTR data is first added, and will initially be linked only to that EUCTR data. After the ClinicalTrials.gov (CTG) record is added the following week, the EUCTR data becomes 'non-preferred', and the CTG ''data'' will be aggregated before the EUCTR data, but the ECRIN ''study Id'' must not change - it must remain as originally allocated. The StudyId allocated to the CTG record must therefore be that generated by the EUCTR record.<br/> | ||
| + | To make that happen, the study_id field of the study_study_links table is first updated using the data in the ''preferred'' side of the table, i.e. preferred source and sd_sid are matched against the study_ids table. This will provide a study_id for all the study pairs in the table where the preferred data has previously been aggregated, whether or not the non-preferred data is new in this aggregation process or not. If however, it is the preferred data that is new, and the non-preferred data that is already referenced in the study_ids table, the study_id must be provided by a second update, where the links table study_id is null, matching the ''non-preferred'' source id and sd_sid to retrieve the study id. Thus, if a study appears to be new and the links table is checked to see if this is the case, (see below) a study Id can be retrieved.<br/> | ||
| + | There is another scenario, where the two (or or more) equivalent identifiers in the links table are ''all'' new to the system, i.e. have been added and appeared in the system within the same week. In that case neither of the updates described above will bring back a study id. This is not a problem for the 'preferred' version of the study data, which will be added first. It will be seen as completely new to the system and be allocated a study Id automatically, as all new studies are. To ensure that the non-preferred duplicated study will always be allocated the correct study_id, however, requires that the study_links table is updated after each addition of new study_ids to the system. Specifically, once all study_ids have been allocated for a source's studies, the links table should again be updated so that any with a null study_id record that match the new 'preferred' study ids can have that study id added to the table.<br/> | ||
| + | Note that for future aggregations the system automatically ensures that the correct id is used. In the example above, for instance, for the the CTG and EUCTR data the study id will be found by a simple matching against the study_ids table. But if another new version of the study data appears, this time in the Peruvian trial registry, the first update described above will match the study links record with the CTG 'preferred' source id and source study identifier, and the Peruvian data will be matched to that study Id as returned from the CTG entry in the study_ids table. But because of what happened the first time this entry appeared, that value will be the one that was originally matched with the ''EUCTR entry''. In other words the study_ids table preserves the original Id allocation, however it was created.<br/> | ||
| + | During the study aggregation process | ||
| + | * The source ids and source study ids are first checked against the study_ids table. Those with a direct match are givenn the study_id from that table. | ||
| + | * If not a direct match, the study may be a new less-preferred or new more-preferred version of an existing study. Both sides of the link table are checked, preferred then non-preferred. If either matches with a source id / source study id pair the ECRIN study id can be obtained directly from the table, and the study ids table updated accordingly. | ||
| + | * if there is still no match the study must be completely new to the system. It will be allocated a new study id using the study_ids table. | ||
| + | * If the new study is part of a linked pair, it will be the preferred side of that pair (by definition, as preferred data is added first) and the preferred source and source study ids can be used to update the study link record with the new study id. | ||
| + | As a separate process, (i.e. independent of the allocation of the study id) the new study id is marked as belonging to a preferred or non-preferred data source. If the data is designated as being from a non-preferred source then, as described above, different rules are applied in adding the data. In the example given, when the EUCTR data is added it will be recognised as being the non-preferred version of the data already aggregated into the system from CTG, so only new or additional data will be added to the study record and its attributes, even though the study id within the MDR is as originally allocated to the EUCTR data.<br/> | ||
===Final Steps=== | ===Final Steps=== | ||
| + | The various temporary tables that remain are removed. Note that at a later stage in the aggregation process, after all study data has been aggregated, the one-to-many linked study data are then added from the nk schema to the study_relationships table in the study (st) schema. | ||
| + | <div style="font-family: monospace; font-size: 13px" > | ||
| + | <pre> | ||
| + | public void AddStudyStudyRelationshipRecords() | ||
| + | { | ||
| + | // Use the study_all_ids to insert the study Ids | ||
| + | // for the linked sources / sd_sids, using | ||
| + | // nk.linked_study_groups as the source | ||
| + | |||
| + | using (var conn = new NpgsqlConnection(connString)) | ||
| + | { | ||
| + | string sql_string = @"Insert into st.study_relationships | ||
| + | (study_id, relationship_type_id, target_study_id) | ||
| + | select s1.study_id, g.relationship_id, s2.study_id | ||
| + | from nk.linked_study_groups g | ||
| + | inner join nk.all_ids_studies s1 | ||
| + | on g.source_id = s1.source_id | ||
| + | and g.sd_sid = s1.sd_sid | ||
| + | inner join nk.all_ids_studies s2 | ||
| + | on g.target_source_id = s2.source_id | ||
| + | and g.target_sd_sid = s2.sd_sid"; | ||
| + | |||
| + | conn.Execute(sql_string); | ||
| + | } | ||
| + | } | ||
| + | } | ||
| + | </pre> | ||
| + | </div> | ||
Latest revision as of 00:46, 14 March 2022
Last updated: 14/03/2022
Contents
Introduction - the problem of duplicates
One of the issues that has to be tackled during aggregation of data from different sources is the fact that the same study can be found, and in tens of thousands of cases is found, in more than one study based source, and that it will have a different identifier in each source. This is mostly because studies can be registered in more than one trial registry, especially when local regulations mandate a registration for any study carried out within a particular country or region. This is the case with the EU, which insists all trials involving medicinal products that are run in the EU must be registered in the EUCTR. About a third of these studies, however, are also registered in other registries, especially Clinicaltrials.gov. In addition, within a data repository, studies will usually be referenced by a local id rather than a pre-existing registry id.
Study titles cannot be relied upon to identify the same study in different source locations. A study title is often expressed slightly differently in different contexts, and cannot in any case be relied upon to be unique (even within the same source). It may be that further research will indicate how titles could be reframed (e.g. to a smaller number of lexemes, expressed in a fixed order) to allow duplicate entries to be discovered using text, but for the moment the only easy way of identifying duplicate studies is by using the 'other identifiers' material found in the source data. Almost all sources contain this material, which includes 'other trial registry' ids, (i.e. other than that used in the source registry entry), as well as ids assigned by the sponsor, funder or, sometimes, a regulatory authority.
These 'other registry ids' can be used to build up a table of study-study links which can then be used during the aggregation process to identify when duplicate studies are being added. In fact the creation of this table is always the first stage of any aggregation. This page describes this process in detail.
This page only describes how duplicate study entries are managed. Duplication between data objects is much rarer, with one exception - the PubMed citations for journal papers. These can also be found multiple times in the source data, both within a single source and across multiple sources. The particular complications in processing PubMed data are described in Processing PubMed Data. The way in which duplications of data objects that are not PubMed citations are managed is described more fully in Aggregating Data.
The Preferred Source concept
If details about a study and its attributes can be found in more than one data source, the obvious question is how should this data be merged in the final aggregated MDR database? In fact there are several aspects to this question:
- How should 'single-occurrence' details about the study be merged? (i.e. the data points that appear once, in the study record itself, such as study display title, study type and status, enrolment target, min and max ages etc.)
- How should study attributes (identifiers, titles, contributors, topics etc.) be merged?
- How should the data objects linked to the same study in different sources (trial registry entries, data and document references) be merged?
- How should data object attributes be merged?
The first question is probably the most difficult. If the system allowed these basic study parameters to be merged from a variety of sources the problem of precedence immediately arises: how could the system 'know' which source to use for each parameter, if they were available in both? For instance how would the system select the enrolment target from one repository rather than another? (There is no guarantee, unfortunately, that the same values will be listed). If the data is edited in one source but not another, should the most recent data always take precedence over the old? (it may be less complete than what is already there).
Because of these issues it was decided to
- Only take the study single occurrence data items from a single data source
- Use the idea of 'preferred sources' to order the precedence of sources in a consistent fashion.
Each data source is therefore given a number that indicate its place in this 'order of preference' - the exact number is not important, only the place it gives to the data source when they are listed, in ascending order, using this parameter. The number is stored within the sf.source_parameters table in the mon database, and is available as a property of the Source objects when they are retrieved from this table. During the aggregation process, the data sources are processed in order, most preferred first, down to the 'least preferred'. In general, more preferred sources have richer or at least more consistent data than less preferred sources. The most preferred source is ClincalTrials.gov, followed (in broad terms) by registries that are extracted individually, then the various trial registries using the WHO dataset, and then data repositories (e.g. Yoda, BioLINCC), and finally object based data sources (PubMed).
Although it cannot be guaranteed in every case, this usually means that the richer data is added to the system first, and that data coming from later, less preferred sources augments rather than replaces it. In general (and as described in more detail in Aggregating Data), during aggregation the core databases are recreated and then completely rebuilt by adding data from each source in turn. If a study does not already exist in the core system when it is 'presented' from a data source database, then it, its attributes, its associated data objects and their attributes are all added during the aggregation process. But if a study is added that already exists in the core system, because it has been added from a 'more preferred' source earlier in the process:
- The single-occurrence study details from the new source are ignored
- Study attributes are only added if they are clearly different from any that have already been added
- Data objects are added (unless, very rarely, they can be seen to be already in the system)
- Data object attributes are added, for all the data object records that are added.
The Preferred Source idea also plays a central role in establishing and using the study-study link data. If a study has multiple source (registry / repository) identifiers in the system, then in general each of those identifiers, other than the 'most preferred', will need to be linked to the 'most preferred' identifier (i.e. the one used by the 'most preferred' source). This is the role of the nk.study_study_links table. When additional 'less preferred' data for a study is added to the system, the first most-preferred instance of the study is guaranteed to already be present in the central mdr study tables, because of the ordering of the addition. The links table is used to match and compare the new data with the existing set, and so determine how the data will be added. How the nk.study_study_links table is constructed is explained in detail below.
Initial Links Data Collection
The initial step is to collect in all the 'other identifier' information from the source databases.
Two temporary tables are established in the links (nk) schema of the mdr database. One simply collects the link data found, and the second is used to hold it once it has been ordered properly.
public void SetUpTempLinkCollectorTable()
{
using (var conn = new NpgsqlConnection(connString))
{
string sql_string = @"DROP TABLE IF EXISTS nk.temp_study_links_collector;
CREATE TABLE nk.temp_study_links_collector(
source_1 int
, sd_sid_1 varchar
, sd_sid_2 varchar
, source_2 int) ";
conn.Execute(sql_string);
}
}
public void SetUpTempLinkSortedTable()
{
using (var conn = new NpgsqlConnection(connString))
{
string sql_string = @"DROP TABLE IF EXISTS nk.temp_study_links_sorted;
CREATE TABLE nk.temp_study_links_sorted(
source_id int
, sd_sid varchar
, preferred_sd_sid varchar
, preferred_source_id int) ";
conn.Execute(sql_string);
}
}
The system then iterates through all the source, interrogating each of the associated databases in turn to bring back the 'other identifier' data, as sd_sid_2, and the id of the organisation assigning the identifier as source_2. That is matched against the source organisation (source_1) and the study's identifier sd_sid (sd_sid_1). The data is pre-filtered to include only identifiers associated with trial registries (and not those assigned from sponsors or funders, etc.). Repository data sources are included in this process but the repository identifiers themselves are not returned - so Yoda and BioLINCC (for example) identifiers appear as sd_sid_1, with a matching sd_sid_2 from a trial registry, but never as sd_sid_2. The data is brought back as an IEnumerable collection of StudyLink objects, which is then immediately stored in the temp_study_links_collector table using the PostgreSQLCopyHelper nuget package, which allows quick and easy storage of batches of data in one call. As the iteration across the sources proceeds, so this table gradually grows with the records found each time.
foreach (Source s in sources)
{
// Fetch the study-study links and store them
// in the Collector table
IEnumerable<StudyLink> links = slh.FetchLinks(s.id, s.database_name);
slh.StoreLinksInTempTable(CopyHelpers.links_helper, links);
}
public IEnumerable<StudyLink> FetchLinks(int source_id, string database_name)
{
string conn_string = repo.GetConnString(database_name);
using (var conn = new NpgsqlConnection(conn_string))
{
string sql_string = @"select " + source_id.ToString() + @" as source_1,
sd_sid as sd_sid_1,
identifier_value as sd_sid_2, identifier_org_id as source_2
from ad.study_identifiers
where identifier_type_id = 11
and identifier_org_id > 100115
and (identifier_org_id < 100133 or identifier_org_id = 101989)
and identifier_org_id <> " + source_id.ToString();
return conn.Query<StudyLink>(sql_string);
}
}
public ulong StoreLinksInTempTable(PostgreSQLCopyHelper<StudyLink> copyHelper, IEnumerable<StudyLink> entities)
{
using (var conn = new NpgsqlConnection(connString))
{
conn.Open();
return copyHelper.SaveAll(conn, entities);
}
}
Cleaning the 'other Ids'
One of the difficulties in the process is that the other identifier data is rarely constrained in the source systems - it can therefore sometimes be poorly formatted or otherwise mis-entered. It is necessary to try and clean some of the data before proceeding further, using a succession of SQL statements - a few of which are shown below. In the examples shown, the SQL a) deletes WHO Universal trial numbers incorrectly entered as trial registry Ids, b) replaces 'n dashes' with ordinary hyphens, making later processing easier, c) updates entries indicating an Australian ACTRN identifier, that are not prefixed by 'ACTRN', and d) removes the redundant 'Chinese Clinical Trial Register' statement from Ids from that registry which include it. These are, however, just 4 examples from a total of over 30 'cleaning' functions.
public void TidyIds1()
{
string sql_string = "";
using (var conn = new NpgsqlConnection(connString))
{
sql_string = @"DELETE from nk.temp_study_links_collector
where sd_sid_2 ilike 'U1111%' or sd_sid_2 ilike 'UTRN%'";
conn.Execute(sql_string);
// replace n dashes
sql_string = @"UPDATE nk.temp_study_links_collector
set sd_sid_2 = replace(sd_sid_2, '–', '-');";
conn.Execute(sql_string);
sql_string = @"UPDATE nk.temp_study_links_collector
SET sd_sid_2 = 'ACTRN' || sd_sid_2
WHERE source_2 = 100116
and length(sd_sid_2) = 14";
conn.Execute(sql_string);
sql_string = @"UPDATE nk.temp_study_links_collector
set sd_sid_2 = replace(sd_sid_2, 'Chinese Clinical Trial Register', '')
where source_2 = 100118;";
conn.Execute(sql_string);
...
...
Once the data is cleaned it needs to be ordered properly, so that preferred and non-preferred ids are clearly identified. This is done by adding the collected data to the nk.temp_study_links_sorted table using two insert statements. The first takes the data that already has the less preferred source/id (i.e. has the higher preference number) as the first two fields, and puts this data straight into the the 'sorted' table. The second takes those where the less preferred source/id are in the last two fields and swaps them round before inserting the data.
public void TransferLinksToSortedTable()
{
using (var conn = new NpgsqlConnection(_connString))
{
// needs to be done twice to keep the ordering of sources correct
// A lower rating means 'more preferred' - i.e. should be used in preference
// Therefore lower rated source data should be in the 'preferred' fields
// and higher rated data should be on the left hand side
// Original data matches what is required
string sql_string = @"INSERT INTO nk.temp_study_links_sorted(
source_id, sd_sid, preferred_sd_sid, preferred_source_id)
SELECT t.source_1, t.sd_sid_1, t.sd_sid_2, t.source_2
FROM nk.temp_study_links_collector t
inner join nk.temp_preferences r1
on t.source_1 = r1.id
inner join nk.temp_preferences r2
on t.source_2 = r2.id
WHERE r1.preference_rating > r2.preference_rating";
int res1 = conn.Execute(sql_string);
// Original data is the opposite of what is required - therefore switch
sql_string = @"INSERT INTO nk.temp_study_links_sorted(
source_id, sd_sid, preferred_sd_sid, preferred_source_id)
SELECT t.source_2, t.sd_sid_2, t.sd_sid_1, t.source_1
FROM nk.temp_study_links_collector t
inner join nk.temp_preferences r1
on t.source_1 = r1.id
inner join nk.temp_preferences r2
on t.source_2 = r2.id
WHERE r1.preference_rating < r2.preference_rating";
int res2 = conn.Execute(sql_string);
_logger.Information((res1 + res2).ToString() + " total study-study links found in source data");
}
}
The sorted table will obviously have many duplicates - data that was entered originally as both source_id_1/sd_sid_1 <=> source_id_2/sd_sid_2, and source_id_2/sd_sid_2 <=> source_id_1/sd_sid_1, will now appear as two identical records: source_id/sd_sid <=> pref_source_id/pref_sd_sid. It is therefore necessary to generate a distinct version of the links data. The nk.temp_distinct_links table is therefore created, containing all the distinct links between studies, with both the more 'preferred' and the less 'preferred' versions of each pair identified. Note that a boolean field called valid is included in the table, with a default value of true. The nk.temp_distinct_links table is used as the maim links table during both the final stages of data cleaning and the next stages of processing.
public void CreateDistinctSourceLinksTable()
{
// The nk.temp_study_links_sorted table will have
// many duplicates... create a distinct version of the data
using (var conn = new NpgsqlConnection(_connString))
{
string sql_string = @"DROP TABLE IF EXISTS nk.temp_distinct_links;
CREATE TABLE nk.temp_distinct_links
as SELECT distinct source_id, sd_sid,
preferred_sd_sid, preferred_source_id, true as valid
FROM nk.temp_study_links_sorted";
conn.Execute(sql_string);
sql_string = @"SELECT COUNT(*) FROM nk.temp_distinct_links";
int res = conn.ExecuteScalar<int>(sql_string);
_logger.Information(res.ToString() + " distinct study-study links found");
}
}
Even though the 'other ids' are now sorted and (mostly) properly formatted they may not all be valid. There may have been errors during data entry, or some other reason why the registry Ids as entered do not correspond to a real study in the system. These invalid ids have to be removed, or the system will try to link data to non-existent studies, causing a variety of problems during the aggregation process. The system therefore loops through each of the sources with study tables, and uses a call to the source database to retrieve a list of the current study ids for that source. These are all guaranteed to be 'real' ids as they have been harvested directly from the sources as the registry ids. They are also the most up to date set of study ids known to the system, being a function of the most recent import processes.
foreach (Source source in sources)
{
...
if (source.has_study_tables)
{
string source_conn_string = _credentials.GetConnectionString(source.database_name, _testing);
slh.ObtainStudyIds(source.id, source_conn_string, CopyHelpers.studyids_checker);
slh.CheckIdsAgainstSourceStudyIds(source.id);
}
}
slh.DeleteInvalidLinks();
public void ObtainStudyIds(int source_id, string source_conn_string, PostgreSQLCopyHelper<IdChecker> copyHelper)
{
using (var conn = new NpgsqlConnection(_connString))
{
string sql_string = @"DROP TABLE IF EXISTS nk.temp_id_checker;
CREATE TABLE nk.temp_id_checker
(sd_sid VARCHAR) ";
conn.Execute(sql_string);
}
IEnumerable<IdChecker> Ids;
using (var conn = new NpgsqlConnection(source_conn_string))
{
string sql_string = @"select sd_sid
from ad.studies";
Ids = conn.Query<IdChecker>(sql_string);
}
using (var conn = new NpgsqlConnection(_connString))
{
conn.Open();
copyHelper.SaveAll(conn, Ids);
}
...
}
Once the study ids have been collected the IDs in the nk.temp_distinct_links table can be checked against them. Both sides (preferred and non-preferred) must be checked separately. If a link contains an invalid study id then it is marked as a invalid. At the end of the process, after all sources have been checked, all the invalid records are deleted. In most cases the proportion of invalid records is very small. In fact only about 50 records are identified (from 32,000 plus) as invalid across all the sources, with one marked exception. That exception is the European Clinical Trials Registry (EU CTR). In this case over 5000 Ids, often called EUDRACT numbers, listed in other trial registries as an 'other registry id', do not correspond to any entry in the EU CTR itself. Checking the relevant titles / key words in several cases indicate that this is not a data entry error issue - the trials simply do not exist in the EU CTR, under any Id. This may be a function of sponsors seeking an EU CTR Id in anticipation of running a study within the EU, or to allow possible expansion to an EU country, but without proceeding to do so.
public void CheckIdsAgainstSourceStudyIds(int source_id)
{
using (var conn = new NpgsqlConnection(_connString))
{
string sql_string = @"UPDATE nk.temp_distinct_links t
SET valid = false
FROM
(SELECT k.sd_sid as sd_sid
FROM nk.temp_distinct_links k
LEFT JOIN nk.temp_id_checker s
ON k.sd_sid = s.sd_sid
WHERE k.source_id = " + source_id.ToString() + @"
AND s.sd_sid is null) invalids
where t.sd_sid = invalids.sd_sid";
int res1 = conn.Execute(sql_string);
_logger.Information(res1.ToString() + " set to invalid on sd_sid");
sql_string = @"UPDATE nk.temp_distinct_links t
SET valid = false
FROM
(SELECT k.preferred_sd_sid
FROM nk.temp_distinct_links k
LEFT JOIN nk.temp_id_checker s
ON k.preferred_sd_sid = s.sd_sid
WHERE k.preferred_source_id = " + source_id.ToString() + @"
AND s.sd_sid is null) invalids
where t.preferred_sd_sid = invalids.preferred_sd_sid";
int res2 = conn.Execute(sql_string);
_logger.Information(res2.ToString() + " set to invalid on preferred_sd_sid");
}
}
The last stage of the process carries out the deletion of the invalid records, drops a temporary table and counts the number of records available for further processing. Importantly it also drops the 'valid' column from the table. That is because it would not be consistentlky updated in later processing, and so would cause the several later calls to 'select distinct' to not function properly. It is therefore simpler to remove it.
public void DeleteInvalidLinks()
{
using (var conn = new NpgsqlConnection(_connString))
{
string sql_string = @"DELETE
FROM nk.temp_distinct_links
WHERE valid = false";
int res = conn.Execute(sql_string);
_logger.Information(res.ToString() + " study-study links deleted because of an invalid study id");
sql_string = @"DROP TABLE IF EXISTS nk.temp_id_checker;";
conn.Execute(sql_string);
sql_string = @"ALTER TABLE nk.temp_distinct_links DROP COLUMN valid;";
conn.Execute(sql_string);
sql_string = @"SELECT COUNT(*) FROM nk.temp_distinct_links";
res = conn.ExecuteScalar<int>(sql_string);
_logger.Information(res.ToString() + " distinct study-study links remaining");
}
}
Exclusion of One-to-Many links
An additional issue is that not all of the links identified between studies are 'simple' one-to-one links. In some cases they are one-to-many. In other words, rather than having the same study listed under different identifiers in different contexts, (scenario 1 in figure 1 below) we have a single study in one source listed as two or more studies in another source (scenario 2). In these circumstances the MDR does not attempt to merge the study data. The system cannot know how data objects and attributes listed on the 'One' side could be allocated to the studies listed on the 'Many' side, so it keeps all studies that are in this sort of relationship as individual studies.
In fact the situation can be more complex - for 30 - 40 groups of studies the relationships are many-to-many. In this case a study may be listed as equivalent to two or more studies in another registry or source, one of which is itself listed as two or more studies (see scenario 3 in figure 1). Usually one of the second set will be the original study, but others are also included. In such case a a group of 4,5 or more (in one case 10!) are listed as equivalent studies in some way. The groupings appear to be a mix of exact equivalents, registered in muiltiple locations, and closely related studies that are listed as equivalent but are not in fact exactly the same (e.g. they may be a follow up study).
Figure 1: Types of linkage between studies
Although the MDR does not attempt to merge the data from one-to-many or many-to-many groupings, it does record the fact that these relationships exist, as a 'study_relationship' record. While some study relationships are between studies listed in the same data source, the ones considered here clearly cut across different sources. The task is therefore to identify this form of linkage and to extract it as study relationship data, removing it from the larger group of 'linked studies'. The first stage is to establish a temporary table that can hold identified study groups, and then identify the groups within nk.temp_distinct_links. A 'group' in this context is two or more studies that are all in the same data source (have the same source id) that are all matched to a particular study. The matching could be at either the preferred or less preferred side of the data so two insert statements are required. Note that at this stage it is just the 'grouping' study and source that are identified (the 'One' side) - not the members of that group (the 'Many' side).
private void Identify1ToNGroupedStudies()
{
string sql_string;
using (var conn = new NpgsqlConnection(_connString))
{
// create a table that takes the rows that involve
// studies with multiple matches in the same source
sql_string = @"DROP TABLE IF EXISTS nk.temp_linked_studies;
CREATE TABLE nk.temp_linked_studies(
group_source_id int
, group_sd_sid varchar
, member_sd_sid varchar
, member_source_id int
, source_side varchar
, complex int DEFAULT 0
)";
conn.Execute(sql_string);
// The source_id side is the group and the preferred side
// is comprised of the grouped studies.
sql_string = @"INSERT into nk.temp_linked_studies
(group_source_id, group_sd_sid,
member_source_id, member_sd_sid,
source_side)
SELECT k.source_id, k.sd_sid,
k.preferred_source_id, k.preferred_sd_sid, 'L'
from nk.temp_distinct_links k
inner join
(SELECT source_id, sd_sid
FROM nk.temp_distinct_links
group by source_id, sd_sid, preferred_source_id
HAVING count(sd_sid) > 1) lhs_groups
ON k.source_id = lhs_groups.source_id
AND k.sd_sid = lhs_groups.sd_sid";
conn.Execute(sql_string);
// The preferred_source_id side is the group and the
// non-prefrred side is comprised of grouped studies
sql_string = @"INSERT into nk.temp_linked_studies
(group_source_id, group_sd_sid,
member_source_id, member_sd_sid,
source_side)
SELECT k.preferred_source_id, k.preferred_sd_sid,
k.source_id, k.sd_sid, 'R' from
nk.temp_distinct_links k
inner join
(SELECT preferred_source_id, preferred_sd_sid
FROM nk.temp_distinct_links
group by preferred_source_id, preferred_sd_sid, source_id
HAVING count(preferred_sd_sid) > 1) rhs_groups
ON k.preferred_source_id = rhs_groups.preferred_source_id
AND k.preferred_sd_sid = rhs_groups.preferred_sd_sid";
conn.Execute(sql_string);
}
}
The temp_linked_studies table now holds all the grouped studies, with the grouping source/studies on the left hand side, and the grouped source/studies on the right. The next stage is to differntiate the studies that belong to the many-to-many relationships, by flagging studies in groups which have members that themselves group other studies. This is done by adding an integer 'complex' flag to the relevant records, first to those involved in the top level group, and then to those in the 'child' group.
private void IdentifyNToNGroupedStudies()
{
string sql_string;
using (var conn = new NpgsqlConnection(_connString))
{
// ...
sql_string = @"Update nk.temp_linked_studies ks1
SET complex = ks1.complex + 2
FROM nk.temp_linked_studies ks2
WHERE ks1.member_source_id = ks2.group_source_id
AND ks1.member_sd_sid = ks2.group_sd_sid
AND ks1.source_side <> ks2.source_side";
conn.Execute(sql_string);
sql_string = @"Update nk.temp_linked_studies ks1
SET complex = ks1.complex + 4
FROM nk.temp_linked_studies ks2
WHERE ks1.group_source_id = ks2.group_source_id
AND ks1.group_sd_sid = ks2.group_sd_sid
AND ks2.complex = 2";
conn.Execute(sql_string);
}
The temp_linked_data can now be used to create linked study group records with the correct relationship codes. For the simpler set of multiple records (complex score = 0) this is straightforward and carried out with the code below. Two complementary pairs of such relationships are possible:
- 25 Includes target as one of a group of non-registered studies: This study includes<the target study>.That study is not registered independently, but instead shares this registry entry with one or more other non-registered studies.
- 26 Non registered but included within a registered study group: This study is registered as <the target study>, along with one or more other studies that share the same registry entry and id.
- 28 Includes target as one of a group of registered studies: This study includes <the target study>, which is registered elsewhere along with one or more other registered studies, forming a group that collectively equates to this study.
- 29 Registered and is included elsewhere in group: This study is also registered, along with one or more other studies that together form an equivalent group, as <the target study>.
BioLINCC (id = 101900) and Yoda (id = 101901) are the only two non registry study sources at the moment, and this is reflected in the SQL below. The relationship codes for these data repositories are different than for the trial registries.
private void Extract1ToNGroupedStudies()
{
string sql_string;
using (var conn = new NpgsqlConnection(_connString))
{
// ...
sql_string = @"INSERT INTO nk.linked_study_groups
(source_id, sd_sid, relationship_id,
target_sd_sid, target_source_id)
select group_source_id, group_sd_sid,
case when group_source_id = 101900
or group_source_id = 101901 then 25
else 28 end,
member_sd_sid, member_source_id
from nk.temp_linked_studies
where complex = 0;";
int res1 = conn.Execute(sql_string);
_logger.Information(res1.ToString() + " relationship records added as part of 1 to n study relationships");
sql_string = @"INSERT INTO nk.linked_study_groups
(source_id, sd_sid, relationship_id,
target_sd_sid, target_source_id)
select member_source_id, member_sd_sid,
case when member_source_id = 101900
or member_source_id = 101901 then 26
else 29 end,
group_sd_sid, group_source_id
from nk.temp_linked_studies
where complex = 0;";
int res2 = conn.Execute(sql_string);
_logger.Information(res2.ToString() + " relationship records added as part of n to 1 study relationships");
}
}
For the 'complex' groups, although only a relatrively small number are involved, the processing required is quite extensive. The initial task is to extract the groups and their members into a uniform format. The code below does that, using a series of union statements to bring together the various grouping studies and their members. (As usual in line comments have been removed for brevity).
private void ExtractNToNGroupedStudyData()
{
string sql_string;
using (var conn = new NpgsqlConnection(_connString))
{
sql_string = @"drop table if exists nk.temp_multi_links;
create table nk.temp_multi_links as
select s1.group_source_id as s1groupsource, s1.group_sd_sid as s1groupsdsid,
s1.member_source_id as s1membersource, s1.member_sd_sid as s1membersdsid,
s1.complex as s1complex,
s2.group_source_id as s2groupsource, s2.group_sd_sid as s2groupsdsid,
s2.member_source_id as s2membersource, s2.member_sd_sid as s2membersdsid,
s2.complex as s2complex
from nk.temp_linked_studies s1
left join
nk.temp_linked_studies s2
on s1.member_sd_sid = s2.group_sd_sid
where s1.complex > 0
order by s1.group_sd_sid, s1.complex desc, s2.complex desc ";
conn.Execute(sql_string);
sql_string = @"drop table if exists nk.temp_list_studies;
create table nk.temp_list_studies as
select * from
(select s1groupsource as src_id, s1groupsdsid as sid_id, s1groupsource as sourceid, s1groupsdsid as sdsid
from nk.temp_multi_links
union
select s1groupsource, s1groupsdsid, s1membersource, s1membersdsid
from nk.temp_multi_links
union
select s1groupsource, s1groupsdsid, s2groupsource, s2groupsdsid
from nk.temp_multi_links
where s2groupsource is not null
union
select s1groupsource, s1groupsdsid, s2membersource, s2membersdsid
from nk.temp_multi_links
where s2groupsource is not null) tot
order by src_id, sid_id, sourceid, sdsid; ";
conn.Execute(sql_string);
}
}
The difficulty is that the source data will have multiple copies of the same or similar groups, but they will be collected under different 'grouping studies'. This is because the original data is read in multiple different ways, so that a different member of the group will be the 'top' grouping study in each case. It is necessary to rationalise the data so that there is only a single set of group records. The code below does this. It tests the members of each group to see if any of them correspond to a previous grouping study. If a match is found all the members of that group are added to the previous study's group rather than being left as an independent group. In that way the original 70+ groups are reduced to 30+. There will be mnany duplicates within each group, so these are removed using a select distinct call in SQL.
private void RationaliseNToNGroupedStudyData()
{
string sql_string;
using (var conn = new NpgsqlConnection(_connString))
{
// First task is to get all entries in related key study lists to the same (first of the group) key study list
sql_string = @"select distinct src_id, sid_id, false from nk.temp_list_studies order by src_id, sid_id";
IEnumerable<ComplexKeyStudy> key_studies = conn.Query<ComplexKeyStudy>(sql_string);
int current_key_src_id = 0, new_key_src_id = 0, current_src_id = 0;
string current_key_sid_id = "", new_key_sid_id = "", current_sid_id = "";
foreach (ComplexKeyStudy ks in key_studies)
{
// ...
current_key_src_id = ks.src_id;
current_key_sid_id = ks.sid_id;
sql_string = @"select sourceid, sdsid
from nk.temp_list_studies
where src_id = " + current_key_src_id.ToString() + @"
and sid_id = '" + current_key_sid_id + @"'
order by sourceid, sdsid";
IEnumerable<ComplexStudy> studies = conn.Query<ComplexStudy>(sql_string);
bool match_found = false;
foreach (ComplexStudy s in studies)
{
if (!match_found) // if still looking
{
current_src_id = s.sourceid;
current_sid_id = s.sdsid;
foreach (ComplexKeyStudy kks in key_studies)
{
if (kks.considered == true) // for the previous 'considered' key studies only...
{
// does the current study entry matrch any of those?
// If so record the details and break out of the loop
if (kks.src_id == current_src_id && kks.sid_id == current_sid_id)
{
match_found = true;
new_key_src_id = kks.src_id;
new_key_sid_id = kks.sid_id;
break;
}
}
}
}
}
if (match_found)
{
// this is a partial or complete repeat of the existing group
// update the table
sql_string = @"UPDATE nk.temp_list_studies
set src_id = " + new_key_src_id.ToString() + @"
, sid_id = '" + new_key_sid_id + @"'
where src_id = " + current_key_src_id.ToString() + @"
and sid_id = '" + current_key_sid_id + "'";
conn.Execute(sql_string);
}
// if no match and is a 'new group' - can be left as it is
ks.considered = true; // signal this key study as 'considered'
}
// Lots of duplicates now so use SQL to selct distinct
sql_string = @"DROP TABLE if exists nk.temp_list_studies2;
CREATE TABLE nk.temp_list_studies2 as
SELECT distinct * from nk.temp_list_studies order by src_id, sid_id, sourceid, sdsid;
DROP TABLE if exists nk.temp_list_studies;
ALTER TABLE nk.temp_list_studies2 rename to temp_list_studies ";
conn.Execute(sql_string);
}
}
It is now relatively straightforward to loop through the rationalised table and create study relationship records. In this case the relationship is type 30: 'Is grouped with, and is the same or similar to <target study>'. Each study has to be given this relationship record with every other member of its group, and then the reverse record, with the role of source and target reversed, is also constructed. A group of 6 related studies will therefore generate 30 relationship records (2 x (5 + 4 + 3 + 2 + 1)).
private void StoreNToNGroupedStudyData()
{
string sql_string;
using (var conn = new NpgsqlConnection(_connString))
{
sql_string = @"select distinct src_id, sid_id from nk.temp_list_studies order by src_id, sid_id";
IEnumerable<ComplexKeyStudy> key_studies = conn.Query<ComplexKeyStudy>(sql_string);
List<ComplexLink> links = new List<ComplexLink>();
foreach (ComplexKeyStudy ks in key_studies)
{
// get the studies related to the current key study
sql_string = @"select sourceid, sdsid
from nk.temp_list_studies
where src_id = " + ks.src_id.ToString() + @"
and sid_id = '" + ks.sid_id + @"'
order by sourceid, sdsid";
List<ComplexStudy> studies = conn.Query<ComplexStudy>(sql_string).AsList<ComplexStudy>();
int studynum = studies.Count;
for (int i = 0; i < studynum - 1; i++)
{
ComplexStudy s = studies[i];
for (int j = i + 1; j < studynum; j++)
{
ComplexStudy t = studies[j];
links.Add(new ComplexLink(s.sourceid, s.sdsid, 30, t.sourceid, t.sdsid));
links.Add(new ComplexLink(t.sourceid, t.sdsid, 30, s.sourceid, s.sdsid));
}
}
}
// store list of relationships, between all possible pairs
conn.Open();
CopyHelpers.complex_links_helper.SaveAll(conn, links);
_logger.Information(links.Count.ToString() + " relationship records added as part of n to n study relationships");
}
}
Finally, it is necessary to remove the grouped study data from the main set of nk.temp_distinct_links, as shown below, and drop the temporary tables that have been used.
private void DeleteGroupedStudyLinkRecords()
{
string sql_string;
using (var conn = new NpgsqlConnection(_connString))
{
sql_string = @"DELETE FROM nk.temp_distinct_links k
USING nk.temp_linked_studies g
WHERE k.source_id = g.group_source_id
and k.sd_sid = g.group_sd_sid
and k.preferred_source_id = g.member_source_id
and g.source_side = 'L';";
int res1 = conn.Execute(sql_string);
sql_string = @"DELETE FROM nk.temp_distinct_links k
USING nk.temp_linked_studies g
WHERE k.preferred_source_id = g.group_source_id
and k.preferred_sd_sid = g.group_sd_sid
and k.source_id = g.member_source_id
and g.source_side = 'R';";
int res2 = conn.Execute(sql_string);
_logger.Information((res1 + res2).ToString() + " study-study links extracted as grouped (1 to n, n to n) records");
sql_string = @"DROP TABLE IF EXISTS nk.temp_grouping_studies;
DROP TABLE IF EXISTS nk.temp_list_studies;
DROP TABLE IF EXISTS nk.temp_multi_links;
DROP TABLE IF EXISTS nk.temp_linked_studies";
conn.Execute(sql_string);
sql_string = @"SELECT COUNT(*) FROM nk.temp_distinct_links";
int res = conn.ExecuteScalar<int>(sql_string);
_logger.Information(res.ToString() + " distinct study-study links remaining");
}
}
Cascading Links
The linked study data requires further processing, however, to ensure that it is complete and can be used for the aggregation process. Two additional problems can arise when a study is found in three or more sources, and need to be resolved. These occur because there is no guarantee that all possible links, or the 'correct' links, as shown in scenario 1 of figure 2, were in the original data.
The first problem is that although the links table currently identifies the more 'preferred' of two linked study ids, it does not necessarily identify the most preferred when there are three or more links. For instance a study could have identifiers in sources A, B and C, where A is less preferred than B, and B less preferred than C. In the source data there are links between A and B, and B and C, but no link exists between A and C, so the data shows A -> B and B -> C. For the aggregation process to work properly the data has to show A -> C and B -> C, i.e. all links should be to the most preferred id. The B -> C link is fine, but the A -> B link has to be 'telescoped' with the 'B -> C' link, and replaced by the resultant A -> C link. This is represented by scenario 2 of figure 2, (though using R, S and T). The system carries out this telescoping process whether or not the A -> C link was originally present in the data. If it was it is duplicated and the duplicate is simply removed. If it was not then the required link record is generated.
The second problem is that to carry out the telescoping behaviour described above requires the intermediate link (B -> C in the example here) to be present. That link may be missing, as in scenario 3 of figure 2, where there is R -> S and R -> T but no S -> T. The system must therefore add these 'missing links' before it can carry out the telescoping process and ensure that all links point to the most preferred version of the study data.
Figure 2: Repairing the linkage between 3 or more studies
The first step in adding the missing intermediate links is to identify studies that have more than one 'preferred' option - these will always link across to more than one preferred source (e.g. R -> S, R -> T) because the grouped studies (linking to the same source / study) have already been removed. This list of studies (in the example all the studies of type 'R') is inserted into temp_studies_with_multiple_links. The process then creates the temp_missing_links table, adding the right hand side preferred fields, by joining this table with the records in the main links table temp_distinct_links, at the same time incorporating the preference ratings of the sources involved. temp_missing_links therefore holds the 'records of interest' from the original links table - all the R -> S, R -> T pairs - together with the preference ratings for all the Rs, Ss and Ts.
public void AddMissingLinks()
{
string sql_string;
using (var conn = new NpgsqlConnection(_connString))
{
// ...
sql_string = @"DROP TABLE IF EXISTS nk.temp_studies_with_multiple_links;
CREATE TABLE nk.temp_studies_with_multiple_links
as SELECT source_id, sd_sid
from nk.temp_distinct_links
group by source_id, sd_sid
having count(distinct preferred_source_id) > 1;";
conn.Execute(sql_string);
sql_string = @"DROP TABLE IF EXISTS nk.temp_missing_links;
CREATE TABLE nk.temp_missing_links as
select k.source_id, r1.preference_rating as source_rating,
k.sd_sid, k.preferred_source_id, r2.preference_rating, k.preferred_sd_sid
from nk.temp_distinct_links k
inner join nk.temp_studies_with_multiple_links m
on k.source_id = m.source_id
and k.sd_sid = m.sd_sid
inner join nk.temp_preferences r1
on k.source_id = r1.id
inner join nk.temp_preferences r2
on k.preferred_source_id = r2.id
order by k.source_id, k.sd_sid, preferred_source_id;";
conn.Execute(sql_string);
...
...
Now, within the same function, a further temp table is created (temp_new_links) to construct the missing links between the E and F studies. This table (temp_new_links) has 6 fields - two initially populated with the source id / sd_sid, to identify the source records in nk.temp_missing_links, and the next pair that have the source / sd_sid pair that has the least preferred rating, i.e. is the study that will need to be on the left hand side in the new link (E in the example above). Place holders are put in for the new_preferred_source and new_preferred_sd_sid fields. Then the table is updated, but this time with the source / sd_sid pair that has the more preferred rating, i.e. is the study that will need to be on the right hand side in the new link (F in the example above) going into the new_preferred_ fields. Thus, if nk.temp_missing_links includes the fields
D-source id D-sd_sid D-preference E-source id E-sd_sid E-preference
D-source id D-sd_sid D-preference F-source id F-sd_sid F-preference
The fields in temp_new_links will initially be:
D-source id D-sd_sid E-source id E-sd_sid 0 '--'
D-source id D-sd_sid E-source id E-sd_sid 0 '--'
and are then updated to:
D-source id D-sd_sid E-source id E-sd_sid F-source F-sd_sid
D-source id D-sd_sid E-source id E-sd_sid F-source F-sd_sid
The four right hand fields now represent the missing (E -> F) links that were required and as such can be inserted into the temp_distinct_links table (as a distinct selection, as in some cases multiple records are created. Of course in some cases the E -> F links may have already existed, as well as the D -> E, D -> F links. The additional record is then redundant, and is removed by making reselecting the link records as a distincct set, but the process has to be carried out on the assumption that the E -> F type links are missing. Finally, the temporary tables are dropped.
public void AddMissingLinks()
{
...
...
sql_string = @"DROP TABLE IF EXISTS nk.temp_new_links;
CREATE TABLE nk.temp_new_links as
select m.source_id, m.sd_sid, m.preferred_source_id as new_source_id,
m.preferred_sd_sid as new_sd_sid, 0 as new_preferred_source, '' as new_preferred_sd_sid from
nk.temp_missing_links m
inner join
(select source_id, sd_sid, min(preference_rating) as min_rating
from nk.temp_missing_links
group by source_id, sd_sid) mins
on m.source_id = mins.source_id
and m.sd_sid = mins.sd_sid
and m.preference_rating <> mins.min_rating
order by source_id, sd_sid;";
conn.Execute(sql_string);
sql_string = @"UPDATE nk.temp_new_links k
SET new_preferred_source = min_set.preferred_source_id
, new_preferred_sd_sid = min_set.preferred_sd_sid
FROM
(select m.* from
nk.temp_missing_links m
INNER JOIN
(select source_id, sd_sid, min(preference_rating) as min_rating
from nk.temp_missing_links
group by source_id, sd_sid) mins
on m.source_id = mins.source_id
and m.sd_sid = mins.sd_sid
and m.preference_rating = mins.min_rating) min_set
WHERE k.source_id = min_set.source_id
AND k.sd_sid = min_set.sd_sid;";
conn.Execute(sql_string);
sql_string = @"INSERT INTO nk.temp_distinct_links
(source_id, sd_sid, preferred_sd_sid, preferred_source_id)
SELECT distinct new_source_id, new_sd_sid, new_preferred_sd_sid, new_preferred_source from
nk.temp_new_links;";
int res = conn.Execute(sql_string);
_logger.Information(res.ToString() + " new study-study links added to complete linkage chains");
// drop the temp tables
sql_string = @"DROP TABLE IF EXISTS nk.temp_missing_links;
DROP TABLE IF EXISTS nk.temp_studies_with_multiple_links;
DROP TABLE IF EXISTS nk.temp_new_links;";
conn.Execute(sql_string);
}
MakeLinksDistinct();
}
Having filled in any 'missing links' the system can safely tackle the telescoping of links. In this scenario, (both A -> B and B -> C exist but not A -> C) the same study (B in this case) will appear in both the left hand side 'less preferred' columns in the temp_distinct_links table and in the more preferred right hand side. The requirement is to replace the B on the right hand side with the 'true' most preferred study id, which is C. A self join on the temp_distinct_links table, linking studies on both 'sides' of the table, can be used to first see if there are any studies that are both 'less' and 'more' preferred, and then to make the switch, replacing the preferred side data on the right of the table with the 'most preferred' values. In the example, A -> B is linked to B -> C by the join, and A -> B becomes A -> C after the update, 'telescoping' the links together.
Because a few studies are registered 4 or even more times the process needs to be repeated until no further studies are found which exist in both the less and more preferred columns. A while loop is therefore used to repeat the action as often as is necessary - usually twice.
In some cases the telescoped link may have already been present - i.e. A -> B, B -> C and A -> C were all there. In these cases the process described above (still necessary to remove the A -> B link) will result in duplicates. Any duplicates therefore have to be removed at the end of the process. This is done by a select distinct. The code below, as usual, has in-line comments reemoved for brevity.
public void CascadeLinks()
{
using (var conn = new NpgsqlConnection(_connString))
{
string sql_string;
int match_number = 500; // arbitrary start number
while (match_number > 0)
{
sql_string = @"SELECT count(*)
FROM nk.temp_distinct_links t1
inner join nk.temp_distinct_links t2
on t1.preferred_source_id = t2.source_id
and t1.preferred_sd_sid = t2.sd_sid";
match_number = conn.ExecuteScalar<int>(sql_string);
_logger.Information(match_number.ToString() + " cascading study-study links found, to 'telescope'");
if (match_number > 0)
{
// do the update
sql_string = @"UPDATE nk.temp_distinct_links t1
SET preferred_source_id = t2.preferred_source_id,
preferred_sd_sid = t2.preferred_sd_sid
FROM nk.temp_distinct_links t2
WHERE t1.preferred_source_id = t2.source_id
AND t1.preferred_sd_sid = t2.sd_sid";
conn.Execute(sql_string);
}
}
}
MakeLinksDistinct();
}
The processing described above, of first adding any missing links and then telescoping the links together, can lead to a small number of new 'groupings' appearing, when more than one study in a single source is linked to a single study elsewhere. The system therefore re-runs the 'exclusion of one-many' routines, to remove these from the nk.temp_distinct_links table.
Updating links with Study_Id
The data is then transferred from the temporary table to the study_study_links table - a table that is created anew at the beginning of each aggregation porcess. Again a select distinct is used, as a final check to ensure that any duplicates are removed.
public void TransferNewLinksToDataTable()
{
// A distinct selection is required because the most recent
// link cascade may have generated duplicates
using (var conn = new NpgsqlConnection(connString))
{
string sql_string = @"Insert into nk.study_study_links
(source_id, sd_sid, preferred_sd_sid, preferred_source_id)
select distinct source_id, sd_sid, preferred_sd_sid, preferred_source_id
from nk.temp_distinct_links";
conn.Execute(sql_string);
}
}
The study links table is then updated with the study Id to be used for each non-preferred / preferred pair, as described below, and is then available to play two critical roles during the aggregation process.
- To ensure that a study Id is allocated consistently across all aggregations, and
- To check if a study is a 'non-preferred' version of a study that has been added earlier, and thus modify the details of the data aggregation process
These two tasks are distinct because a study's non-preferred entry may be added to the MDR before its preferred set of data, for example the EU CTR entry may be added a week before the same study appears in ClinicalTrials.gov. In that example, the persistent study Id within the system (the ECRIN ID) will be allocated when the EUCTR data is first added, and will initially be linked only to that EUCTR data. After the ClinicalTrials.gov (CTG) record is added the following week, the EUCTR data becomes 'non-preferred', and the CTG data will be aggregated before the EUCTR data, but the ECRIN study Id must not change - it must remain as originally allocated. The StudyId allocated to the CTG record must therefore be that generated by the EUCTR record.
To make that happen, the study_id field of the study_study_links table is first updated using the data in the preferred side of the table, i.e. preferred source and sd_sid are matched against the study_ids table. This will provide a study_id for all the study pairs in the table where the preferred data has previously been aggregated, whether or not the non-preferred data is new in this aggregation process or not. If however, it is the preferred data that is new, and the non-preferred data that is already referenced in the study_ids table, the study_id must be provided by a second update, where the links table study_id is null, matching the non-preferred source id and sd_sid to retrieve the study id. Thus, if a study appears to be new and the links table is checked to see if this is the case, (see below) a study Id can be retrieved.
There is another scenario, where the two (or or more) equivalent identifiers in the links table are all new to the system, i.e. have been added and appeared in the system within the same week. In that case neither of the updates described above will bring back a study id. This is not a problem for the 'preferred' version of the study data, which will be added first. It will be seen as completely new to the system and be allocated a study Id automatically, as all new studies are. To ensure that the non-preferred duplicated study will always be allocated the correct study_id, however, requires that the study_links table is updated after each addition of new study_ids to the system. Specifically, once all study_ids have been allocated for a source's studies, the links table should again be updated so that any with a null study_id record that match the new 'preferred' study ids can have that study id added to the table.
Note that for future aggregations the system automatically ensures that the correct id is used. In the example above, for instance, for the the CTG and EUCTR data the study id will be found by a simple matching against the study_ids table. But if another new version of the study data appears, this time in the Peruvian trial registry, the first update described above will match the study links record with the CTG 'preferred' source id and source study identifier, and the Peruvian data will be matched to that study Id as returned from the CTG entry in the study_ids table. But because of what happened the first time this entry appeared, that value will be the one that was originally matched with the EUCTR entry. In other words the study_ids table preserves the original Id allocation, however it was created.
During the study aggregation process
- The source ids and source study ids are first checked against the study_ids table. Those with a direct match are givenn the study_id from that table.
- If not a direct match, the study may be a new less-preferred or new more-preferred version of an existing study. Both sides of the link table are checked, preferred then non-preferred. If either matches with a source id / source study id pair the ECRIN study id can be obtained directly from the table, and the study ids table updated accordingly.
- if there is still no match the study must be completely new to the system. It will be allocated a new study id using the study_ids table.
- If the new study is part of a linked pair, it will be the preferred side of that pair (by definition, as preferred data is added first) and the preferred source and source study ids can be used to update the study link record with the new study id.
As a separate process, (i.e. independent of the allocation of the study id) the new study id is marked as belonging to a preferred or non-preferred data source. If the data is designated as being from a non-preferred source then, as described above, different rules are applied in adding the data. In the example given, when the EUCTR data is added it will be recognised as being the non-preferred version of the data already aggregated into the system from CTG, so only new or additional data will be added to the study record and its attributes, even though the study id within the MDR is as originally allocated to the EUCTR data.
Final Steps
The various temporary tables that remain are removed. Note that at a later stage in the aggregation process, after all study data has been aggregated, the one-to-many linked study data are then added from the nk schema to the study_relationships table in the study (st) schema.
public void AddStudyStudyRelationshipRecords()
{
// Use the study_all_ids to insert the study Ids
// for the linked sources / sd_sids, using
// nk.linked_study_groups as the source
using (var conn = new NpgsqlConnection(connString))
{
string sql_string = @"Insert into st.study_relationships
(study_id, relationship_type_id, target_study_id)
select s1.study_id, g.relationship_id, s2.study_id
from nk.linked_study_groups g
inner join nk.all_ids_studies s1
on g.source_id = s1.source_id
and g.sd_sid = s1.sd_sid
inner join nk.all_ids_studies s2
on g.target_source_id = s2.source_id
and g.target_sd_sid = s2.sd_sid";
conn.Execute(sql_string);
}
}
}