Missing PIDs and Hashing
Last updated: 19/05/2022
Contents
Identifiers and Issues
All records within the MDR system are identified by using Id fields, with relationships between records represented by the use of some of those Id fields as foreign keys. Within the sd and ad tables in the 'source' databases (i.e. the ones that hold the data for a single data source), all tables are constructed with an integer identity key, that is generated automatically by the system in ascending sequence. In addition study tables have a 'source data study identifier' field: sd_sid, and data object tables have a 'source data object identifier field: sd_oid, as shown in the excerpts from the table creation code below.
CREATE TABLE sd.studies(
id INT GENERATED ALWAYS AS IDENTITY PRIMARY KEY
, sd_sid VARCHAR NOT NULL
, display_title VARCHAR NULL
, title_lang_code VARCHAR NULL default 'en'
, brief_description VARCHAR NULL
, ...
, ...
CREATE TABLE ad.study_identifiers(
id INT GENERATED ALWAYS AS IDENTITY PRIMARY KEY
, sd_sid VARCHAR NOT NULL
, identifier_value VARCHAR NULL
, ...
, ...
CREATE TABLE ad.data_objects(
id INT GENERATED ALWAYS AS IDENTITY PRIMARY KEY
, sd_oid VARCHAR NOT NULL
, sd_sid VARCHAR NULL
, title VARCHAR NULL
, ...
, ...
CREATE TABLE ad.object_dates(
id INT GENERATED ALWAYS AS IDENTITY PRIMARY KEY
, sd_oid VARCHAR NULL
, date_type_id INT NULL
, ...
, ...
The integer Id fields are not persistent (for example in the sd schemas they are destroyed and recreated during each harvest) and are not designed to be used outside of their immediate context. They are there as part of normal database 'good practice', to allow records to be uniquely identified, e.g. for editing in pgAdmin table browser, and play little direct part in the processes within the MDR. The sd_sid and sd_oid fields, however, are crucial. These are derived from the source data in fixed ways, and can usually be regarded as persistent, though there are a few problems with their derivation, as described below. They are used, as foreign keys, to bind both attributes and data objects to the 'source' study, as well as data object attributes to the object itself. sd_sids and sd_oids are unique within the studies or data objects tables in any source database, but they cannot be guaranteed to be unique across the whole of the MDR, even though in practice they often are. To uniquely identify a study or object across the MDR the sd_sid or sd_oid must be combined with the source id (a 6 digit identifier). In the individual source databases the addition of the source id is not required, but in the mdr database itself, that contains data from all sources, the combination of source id and sd_sid / sd_oid is neeeded.
In the mdr database, where all data is aggregated, the studies and data objects do receive persistent, unique integer Ids, which can be interpreted as 'ECRIN Accession Ids'. These can be equated to one or more sd_sids or sd_oids because studies, and objects, may be present in one or more sources. The mdr database includes tables that store the relationship between the source data identifiers and the matching ECRIN Ids.
CREATE TABLE core.studies(
id INT NOT NULL PRIMARY KEY
, display_title VARCHAR NULL
, title_lang_code VARCHAR NULL
, ...
, ...
CREATE TABLE core.study_identifiers(
id INT NOT NULL PRIMARY KEY
, study_id INT NOT NULL
, identifier_value VARCHAR NULL
, ...
, ...
CREATE TABLE core.data_objects(
id INT NOT NULL PRIMARY KEY
, display_title VARCHAR NULL
, ...
, ...
CREATE TABLE core.object_dates(
id INT NOT NULL PRIMARY KEY
, object_id INT NOT NULL
, date_type_id INT NULL
, ...
, ...
Note that in the core schema, study and object attributes are grouped with the main study or object by using the study_id or object_id fields, which are the same as the id fields in the study and data object tables respectively. This is similar to the use of sd_sid and sd_oid in the source databases. The link between studies and data objects, however, which in the source database uses sd_sid as a foreign key, no longer exists in the main mdr database. This is because here the links between studies and objects are many to many, whereas in the source databases they are always interpreted as one-to-many. The core schema therefore requires a join table (study_object_links) to resolve the relationship. The way in which source data ids are matched to one another is described more fully in Identifying Links between Studies, whilst the aggregation process, including the way in which the persistent ECRIN Ids are created and maintained, is described in Aggregating Data.
For this scheme to work there are three Id related issues that need to be resolved
a) the lack of globally consistent or guaranteed identifiers for studies, at least for some sources.
b) the lack of simple persistent identifiers (PIDs) in the source data, for most data objects, and
c) the total lack of any identifiers for both study and object attributes.
Each of these problems, and the mechanisms used by the MDR system to resolve them, are described more fully below.
The Study Ids Problem
Most study data in the system is drawn from trial registries. Studies in a registry always have a persistent identifier, e.g. 'NCT05364619', or 'ISRCTN02672532', and this identifier can be used as the source data study identifier, or sd_sid. For all these studies, which represent 99%+ of those in the MDR, the sd_sid is therefore not a problem. For clinical trial repositories, however, such as BioLINCC or Yoda, things are not so simple. Even though the material in the repositories is organised by study, there is not always a persistent identifier available for those studies. BioLINCC, for example, has an accession id for each study but - for reasons that are not clear - in some cases this can change (albeit infrequently). Yoda has no explicit identifier for studies at all (the web page reference here, as in any other system, cannot be guaranteed to remain fixed).
For these non-registry sources it is therefore necessary to create an automatic method for creating sd_sid. This has to be done on a source by source basis. For BioLINCC things are straightforward - each study has an acronym ('TOPCAT', 'ACRN-LARGE') and these do appear to be fixed. It therefore makes sense to simply use the acronym as the sd_sid.
For Yoda things are more difficult. There is no alpha-numeric code attached to studies and using the title is impossible because in a few cases studies share exactly the same title. Most of the studies in YODA are also registered elsewhere, however, in almost every case in Clinicaltrials.gov, and they therefore have an associated NCT number. When this is the case a Yoda sd_sid can be constructed by simply taking the NCT number and prefixing it with 'Y-', e.g.'Y-NCT00487539'. For about 30 studies listed in Yoda (about 6%) there is no corresponding NCT entry. In these cases the sponsor's Id, usually a relatively short code, can be used instead, again prefixed by 'Y-', e.g. 'Y-GAL-MVD-302'. Although a sponsor's Id also cannot be guaranteed to be present or unique, so far all Yoda studies have either had a trial registty Id or, failing that, a unique sponsor id, and so have always been able to be assigned a straightforward sd_sid. This cannot be absolutely guaranteed in the future, so as an additional 'fall back', if neither registry id or protocol id are present, the system will construct an sd_sid by applying a hash function (MD5) to a concatenation of the study name and the enrolment number.
For the TSD repository, to be used by ECRIN for COVID-19 research, the expectation is that most of the studies in the system will be registered, and that registry id will be known to the Repository Management System. In that case the assigned sd_sid will be the registry id prefixed by TSD, e.g. 'TSD-EU 2020-003698-24' for a trial registered in the EU CTR. If a study is not registered it may be possible to use an Id generated by the TSD system, prefixed by 'TSD', if this can be guaranteed to be persistent and available - this currently (May 2022) needs to be confirmed. The fall back mechanism would again involve an MD5 hash of a small subset of study data - e.g. title, type code, start year, start month, enrolment number.
As other non registry sources are added to the system it will be necessary to construct similar mechanisms for consistent generation of sd_sids for each. Unfortunately this will have to be done on a source by source basis.
The Data Object Ids Problem
Most but not all data objects in the system have an associated URL, but that link cannot be guaranteed to be permanent. It is also true that journal articles have a Pubmed id and / or a doi, but journal articles are handled by the system differently from how most other data objects are processed. They are linked to studies after extraction, while most other objects are linked, if only implicitly, to studies in the sources (trial registries, data repositories) where they are found during extraction.
In general, the MDR system therefore has to construct an identifier for each data object, using the source data in a consistent way to guarantee that the same sd_oids are created each time the data is examined. In the great majority of cases the data objects associated with a study will all be of different types - e.g. a trial registry entry, a results summary in a trial registry, a protocol, a dataset, a statistical analysis plan. It is therefore possible - at least in most cases - to use the combination of the sd_sid, to uniquely identify the source study, the type code, and the object name (if it has one) or type (if it has no name) to construct a source data object identifier for any data object. The MDR does exactly this as each data object is identified, and the resultant sd_oid is used when populating the sd object tables in the data source databases, with the system placing a double colon between each component for greater legibility. Examples are:
NCT04397159 :: 13 :: CTG registry entry
BABY HUG :: 35 :: BABY HUG Follow-up Study II Manual of Operations
NCT04537910 :: 22 :: Statistical Analysis Plan
Y-NCT00207766 :: 79 :: CSR Summary
ISRCTN15279830 :: 11 :: 40046_PROTOCOL_V5.0_23Jul21.pdf
There are situations where a study may be associated with more than one object of the same type, e.g. for different substudies, as in the second example above, or as different versions of the same document, as in the 5th example. In almost all of these cases the object will have a different name, or label, so the sd_oid mechanism still produces a unique identifier. In a relatively few cases the extraction mechanism may not differentiate the files fully, for instance objects labelled the same in the source data but with a different file name, so giving the object the same sd_oid within the system. In practice this mostly occurs in the context of identifying documents in some sections of the ClinicalTrials.gov XML file record - for example in sections headed 'See also', or 'Large documents'. To help ensure unique sd_oids the extraction mechanism in these contexts checks whether the name proposed for the object has been used before. If it has a suffix (_1 for the second object, _2 for the third, and so on) is added to the name, as illustrated below...
// title type depends on whether label is present
if (!string.IsNullOrEmpty(doc_label))
{
object_display_title = title_base + " :: " + doc_label;
t_type_id = 21; t_type = "Study short name :: object name";
}
else
{
object_display_title = title_base + " :: " + object_type;
t_type_id = 22; t_type = "Study short name :: object type";
}
// check name
int next_num = CheckObjectName(object_titles, object_display_title);
if (next_num > 0)
{
object_display_title += "_" + next_num.ToString();
}
// title is right hand part of full display title, after ' :: '
object_title = object_display_title.Substring(title_base.Length + 4);
sd_oid = sid + " :: " + object_type_id.ToString() + " :: " + object_title;
...
...
// check name...
private int CheckObjectName(List<ObjectTitle> titles, string object_display_title)
{
int num_of_this_type = 0;
if (titles.Count > 0)
{
for (int j = 0; j < titles.Count; j++)
{
if (titles[j].title_text.Contains(object_display_title))
{
num_of_this_type++;
}
}
}
return num_of_this_type;
}
The addition of a suffix in this way is still a relatively rare event. Within the 500,000+ data objects listed for the studies in Clinicaltrials.gov, only 88 have a _1 suffix, 6 have _2, and 3 have _3.
The exception to the sd_oid derivation mechanism described above relates to published papers. A study will very often have several papers associated with it, but each of these will have their own identifier. This is because all the papers known to the MDR, at least at the moment, have a PubMed Id. This is an integer, though for compatibility with other sd_oids it is imported into ther MDR as a string.
Taken together, the mechanisms described in thbis section provide a unique sd_oid for each data object in the system.
The Study / Object Attribute Ids Problem
Study and object attributes - identifiers, titles, instances, dates, topics etc. - have no associated intrinsic identifiers at all. A significant additional problem, especially when web scraping is used, is that there is no guarantee that attributes will be processed in the same order as before, or will be present in the same numbers, which means the system has to compare one unordered set of records with another to identify the new, edited and (more rarely but not impossible) deleted items.
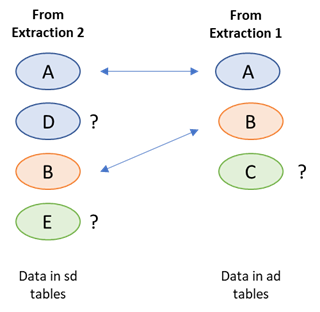
Imagine a study with 3 identifier attributes a, b and c. a is a registry identifier and b and c are sponsor ids. On the next download a funder identifier entry ‘d’ is found, which is the second in the list, while at the same time identifer c has been edited, to become 'e' – the list is now a, d, b and e. The two sets of study identifiers can be compared and a match can be found between the two versions of a and b. But d and e are new to the system and c can no longer be matched. One of d or e corresponds to the pre-edited version of c (see figure 1), but which one?
Records which are identical in each set can be matched, but for objects that are not matched
it is impossible, in general, to determine their origin or relationship with objects in the previous set.
A human could look at the data and, in most cases, seeing that c and e are ‘similar’ can deduce that c has been edited into e, and d is completely new. But a machine cannot do that without a complex scoring / weighting algorithm, that would need to take into account every field in the record, and even then could only make an estimate rather than being 100% certain (the more extensive the editing, the closer the record that results looks like a completely new addition). The result is that the system has to
a) Mark both d and e as new attributes and add them both to the system
b) Mark c as no longer relevant, and ‘retire’ it from the system.
In other words edits have to be handled explicitly as a combination of insert and delete.
It is true that for some types of attribute sets (e.g. object dates), you could make assumptions about the data and say, for example, that a data object would only ever have one date of a certain type. But this mechanism is only possible for some types of attribute (for example studies may have multiple identifiers of the same type) and assumptions about uniqueness of type are dangerous to make with very large data sets, often with poorly managed data. The need is therefore to find an efficient method for identifying changes in sets of attributes, and for actioning the changes that are required.
The general answer is to use a hashing technique to encapsulate the key fields of any record into a single field. The hashes that result can be used to efficently compare both the main entities (studies and data objects), and related groups of study and object attributes, between successive data imports. As described below, this removes the need for identifiers for attribute records altogether.
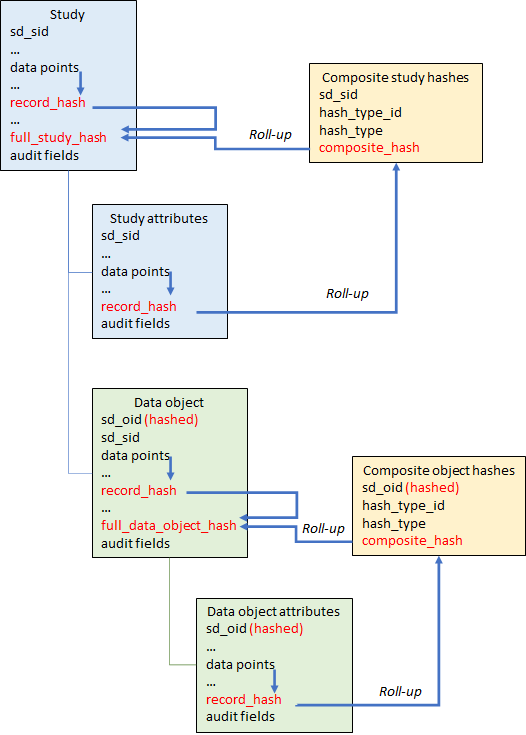
It is first necessary to be clear about what ‘comparing’ two records means, and how equality (or its converse, an edit) of records is defined. The system could compare every field in each record, but in fact only some fields are relevant to the record’s 'essential content' and need to be included in the comparison. Id fields, whether from the source data or generated as described above, a foreign id that link an attribute record to its parent entity, and audit fields that date the record, do not need to be compared, just the fields that make up the record's 'data payload' (see figure 2). In addition, because the ad records only have the codes for categorised questions, and not the text decode (whereas the sd equivalent tables have both), only the code needs to be included in the sd hashes used for comparison purposes.
If an id is not available or not constant, then the payload fields need to be used to identify and match the record.
Using hashes means that rather than comparing each field in turn, a process that would be different for each record type, it is easier to first ‘roll up’ the relevant values into a single string and then compare only that field. If the values are equal the records are equal, in the sense that their content-containing fields are the same. If they are unequal there is a difference in one or more of those fields, and the new record needs to replace the old. Each table in the sd and ad schemas therefore includes a hash_record field. Using the md5 algorithm, a string of 32 hex characters (=128 bits) is created whatever the length of the source string. The hash value is almost certain to be unique – there are 2^128 or 3.4 X 10^38 possible values (using the text itself could result in very long strings that would take much longer to compare).
This process is most easily carried out in postgres itself, first using the json_build_array function to turn a subset of field values in a record into a json array, then casting that array into a string, and then hashing that string using the built in md5 function. For instance, to produce a hash for the study identifiers records and store it in a field called record_hash, the command below can be used:
public void create_study_identifier_hashes()
{
string sql_string = @"Update sd.study_identifiers
set record_hash = md5(json_build_array(identifier_value, identifier_type_id, identifier_org_id,
identifier_org, identifier_date, identifier_link)::varchar)";
h.ExecuteHashSQL(sql_string, "study_identifiers");
}
This provides a very simple way of creating a summary record_hash field for every record in the sd tables, later to be transferred into the ad tables. The main difficulty is that hashing is computationally expensive and with very large tables the system sometimes 'times out'. Along with the need to provide some periodic feedback to the calling program and / or user, it is therefore useful to split up the hashing process. Hence the call to ExecuteHashSQL. This function - for very large tables - splits the records to be hashed up into batches of 500,000, and constructs the freedback message(s) accordingly. It does so by adding an additional where condition to the sql string passed into it. The function is shown below...
public void ExecuteHashSQL(string sql_string, string table_name)
{
try
{
int rec_count = GetRecordCount(table_name);
int rec_batch = 500000;
// int rec_batch = 10000; // for testing
if (rec_count > rec_batch)
{
for (int r = 1; r <= rec_count; r += rec_batch)
{
string batch_sql_string = sql_string + " where id >= " + r.ToString() + " and id < " + (r + rec_batch).ToString();
ExecuteSQL(batch_sql_string);
string feedback = "Creating " + table_name + " hash codes, " + r.ToString() + " to ";
feedback += (r + rec_batch < rec_count) ? (r + rec_batch - 1).ToString() : rec_count.ToString();
StringHelpers.SendFeedback(feedback);
}
}
else
{
ExecuteSQL(sql_string);
StringHelpers.SendFeedback("Creating " + table_name + " hash codes - as a single batch");
}
}
catch (Exception e)
{
string res = e.Message;
StringHelpers.SendError("In hash creation (" + table_name + "): " + res);
}
}
In addition, for a group of related records, the hash values can themselves be rolled up to form a hash that corresponds to the total content of that group. This allows a group of records to be easily compared. For example, to create a hash for the whole group of study identifiers listed for each study, the following code can be used (sd_sid is the source data study id, usually the registry id, the hash_type_id and hash type are 11 and 'identifiers' respectively, in this instance, and the table_name would be 'study_identifiers'):
public void create_composite_study_hashes(int hash_type_id, string hash_type, string table_name)
{
string top_sql_string = @"Insert into sd.study_hashes
(sd_sid, hash_type_id, hash_type, composite_hash)
select t.sd_sid, " + hash_type_id.ToString() + ", '" + hash_type + @"',
md5(to_json(array_agg(t.record_hash ORDER BY t.record_hash))::varchar)
from sd." + table_name;
h.CreateCompositeStudyHashes(top_sql_string, hash_type_id, hash_type);
}
This function can be called for all study attribute types by changing the parameters, and a similar function exists for data object attribute tables. Again it is necessary to split up very large tables because the process is time consuming - the CreateCompositeStudyHashes function is shown below, and again uses the ids in the source to split up records into batches (this time of only 10,000) where necessary.
public void CreateCompositeStudyHashes(string top_sql_string, int hash_type_id, string hash_type)
{
try
{
int rec_count = GetRecordCount("studies");
int rec_batch = 10000;
//int rec_batch = 1000; // for testing
if (rec_count > rec_batch)
{
for (int r = 1; r <= rec_count; r += rec_batch)
{
string where_sql_string = " where s.id >= " + r.ToString() + " and s.id < " + (r + rec_batch).ToString();
string batch_sql_string = top_sql_string + @" t
inner join sd.studies s
on s.sd_sid = t.sd_sid
" + where_sql_string + " group by t.sd_sid;";
ExecuteSQL(batch_sql_string);
string feedback = "Creating composite study hash codes (" + hash_type + "), " + r.ToString() + " to ";
feedback += (r + rec_batch < rec_count) ? (r + rec_batch - 1).ToString() : rec_count.ToString();
StringHelpers.SendFeedback(feedback);
}
}
else
{
string sql_string = top_sql_string + @" t group by t.sd_sid;";
ExecuteSQL(sql_string);
StringHelpers.SendFeedback("Creating composite study hash codes (" + hash_type + ") as a single batch");
}
}
catch (Exception e)
{
string res = e.Message;
StringHelpers.SendError("In study composite hash creation: " + res);
}
}
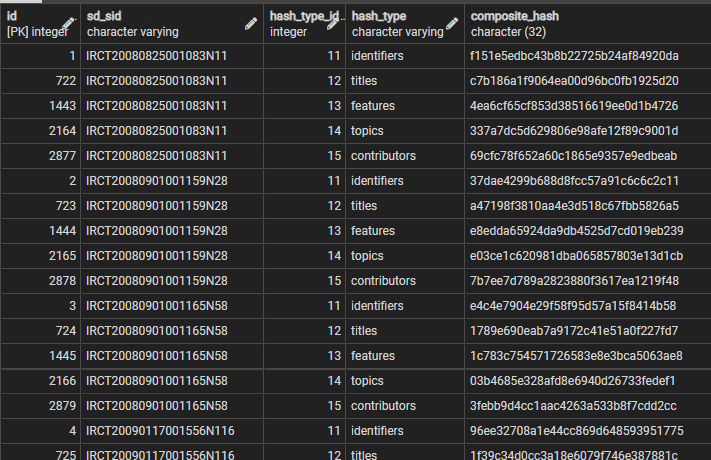
The table that results is illustrated below, for the first few studies listed in the Iranian Trial Registry
With this hash it is now possible to use a single comparison to see if any of the identifier data for a study has changed. The same is true of all other attributes, for studies and data objects. The process can be extended once more, to roll up all of the composite hashes for each study or data object, plus the record hash for the study or data object record itself, to produce a 'full_hash' for the whole entity. Comparing these full hashes allows an edit in any component of the study or data object data to be detected, because the full hash only remains the same if no changes at all have taken place in the 'essential data' carried by each record. If the full hashes are different, then the entity record hashes can be compared, and then the attribute composite hashes, to see where the edit(s) have occurred. This multiple hash roll-up process is shown in figure 4.
– but hashes are also constructed for whole records and used to detect changes in those records, and groups of related records, and finally in the entity (study or data object) as a whole.
In summary, at the end of each data extraction for each source, the system
- Creates a hash equivalent of the (non-id, non-audit) content of all study, study attribute, data object and data object attribute records, in all tables. In all cases this hash is stored in a field called ‘record_hash'.
- Creates a hash for each group of multiple attributes, e.g. for each study – a hash representing all identifiers, a hash for all titles, a hash for all study features, etc. For each data object: a hash for all titles, a hash for all instances, a hash for all dates, etc.
- The ‘composite hashes’ are stored in separate tables (one for study hashes, the other for data object hashes) that reference the parent study or data object, indicate the type of composite hash, and the hash value itself.
- The composite hash values for each study are rolled up with the hash for the study record itself to form a composite hash for the study and all of its components, a ‘study full hash’. It is stored in the Studies table.
- Similarly, the composite hash values for each data object are rolled up with the hash for the data object itself to form a composite hash for the data object and all of its components, a ‘data object full hash’. It is stored in the data objects table.
All of the hashes need to be transferred with the data records from the sd (session data) tables to the ad (accumulated data) tables whenever that transfer takes place, i.e. after the comparison process has identified the new or revised data. The details of the way in which these has values are compared and used in the import process is described in more detail on the Importing Data page, but the principle is very siomple - if a difference in the composite hash for a group of attributes, for the same study or object, indicates that a change of any kind has occured, then all of those attributes are deleted in the destination table, to be replaced by the entire set of attributes in the source table. No attempt is made to compare individual records, and therefore attribute records do not need identifiers to distinguish them, beyond the id that links them to their parent entity - a study or data object.